I confirm that all authors on this abstract approve this submission.
Alex Peterson, , Carly TestUser
Login to Planstone with admin link (username/password):
1. Click on "Abstracts" tab
2. Click on "Testusers" tab to the upper right hand side
3. Create a new testuser account and just add your email address to it.
We ask that you test with "Test Accounts" so as not to compromise your admin authority with your regular account.
4. Test through the submission process to provide screenshots/edits of items that need to be edited, if applicable.
Posted Wednesday at 4:20pm
CORONA STUDY: DELTAREX-G, DISEASE-TARGETED GENE THERAPY, FOR COVID-19
Erlinda Gordon, MD, Cancer Center of Southern California/Sar1, Sant Chawla, MD, , Frederick Hall, PhD
Cancer Center of Southern California/Sar1
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This aggressive and highly infectious virus can cause life threatening complications including cytokine storm and acute respiratory distress syndrome. However, these complications are believed to be caused not by the virus itself, but by an exaggerated immune reaction to the infection. The CORONA STUDY is a Phase 1/2 study that will test whether DeltaRex-G is safe, will prevent/reduce the severity of cytokine storm and ARDS, and hasten clinical recovery from COVID-19.
Rationale and Guiding Hypotheses:
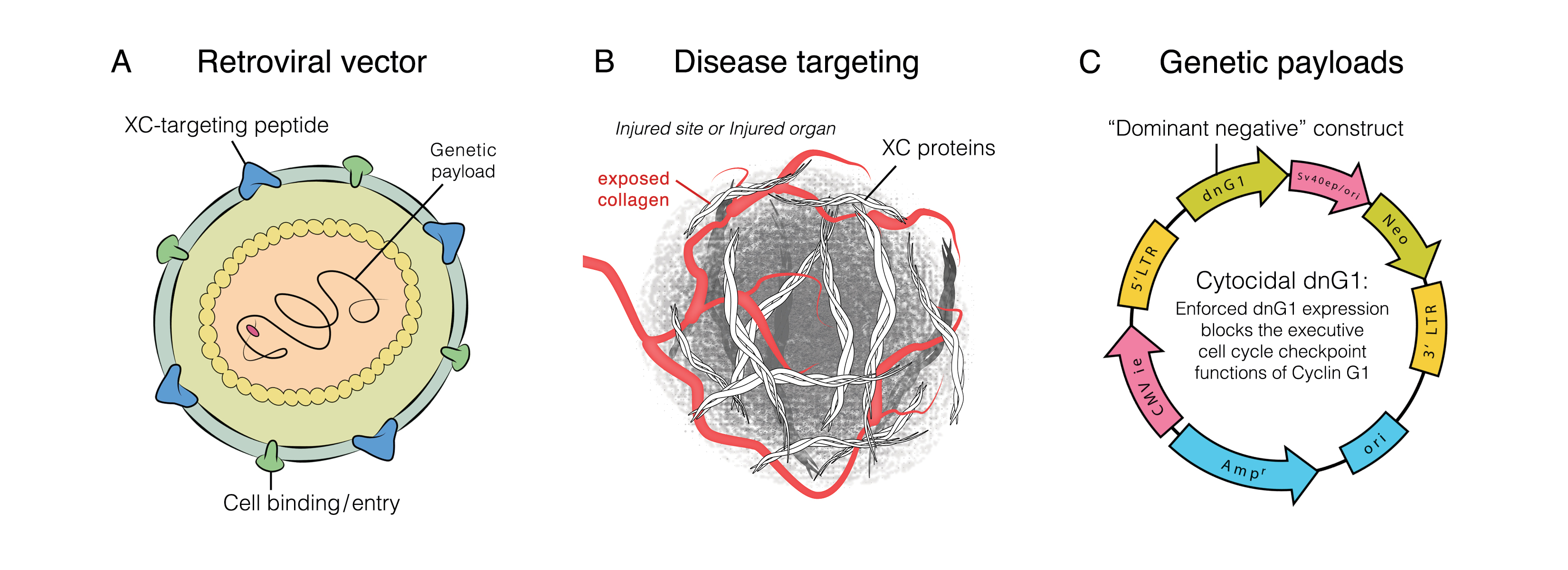
1) DeltaRex-G is a disease-targeted replication-incompetent, amphotropic MLV-based retroviral vector that (a) displays a collagen-matrix binding peptide on its surface for targeting areas of pathology, and (b) encodes a cytocidal dominant negative human cyclin G1 construct. When injected intravenously, the DeltaRex-G nanoparticles seek out and accumulate in areas of injury/pathology where collagen is exposed, in the vicinity of proliferative immune cells. DeltaRex-G may then enter the rapidly dividing T cells and activated macrophages and would kill them by blocking the G1 phase of the cell division cycle, hence, reducing cytokine release and the severity of cytokine storm and ARDS;
2) Five Phase 1 and 2 US-based clinical trials for metastatic cancer have shown that intravenous DeltaRex-G has minimal systemic toxicity, presumably, due to its navigational system that limits the biodistribution of DeltaRex-G to areas of pathology where exposed collagenous (XC) proteins are abnormally found; and
3) DeltaRex-G is currently available in FDA approved "Right to Try" or Expanded Access Program for Stage 4 cancers (IND# 19130). The dose of DeltaRex-G given to this vulnerable cancer population exhibiting symptomatology similar to COVID-19, is the highest proposed dose for the CORONA study.
A Phase 1/2 clinical trial for CORONA Study: DeltaRex-G for Symptomatic COVID-19 was submitted under IND 22477 and reviewed by the USFDA in July, 2020. Currently, toxicity/biodistribution studies are planned to test the feasibility of using DeltaRex-G in a SARS-CoV-2 susceptible animal model as required by the USFDA. Upon completion of these non-clinical studies, the CORONA study will test whether DeltaRex-G is safe, will improve the symptoms of moderately severe to severe COVID-19, improve cytokine pattern, and will hasten patient recovery from COVID-19.
Plasmapheresis, Anti-ACE2 and Anti-FcγRII Monoclonal antibodies; a possible treatment for severe cases of COVID-19
Amin Sedokani, MD, Urmia University of Medical Sciences1, Sadegh Feizollahzadeh, PhD
Urmia University of Medical Sciences1
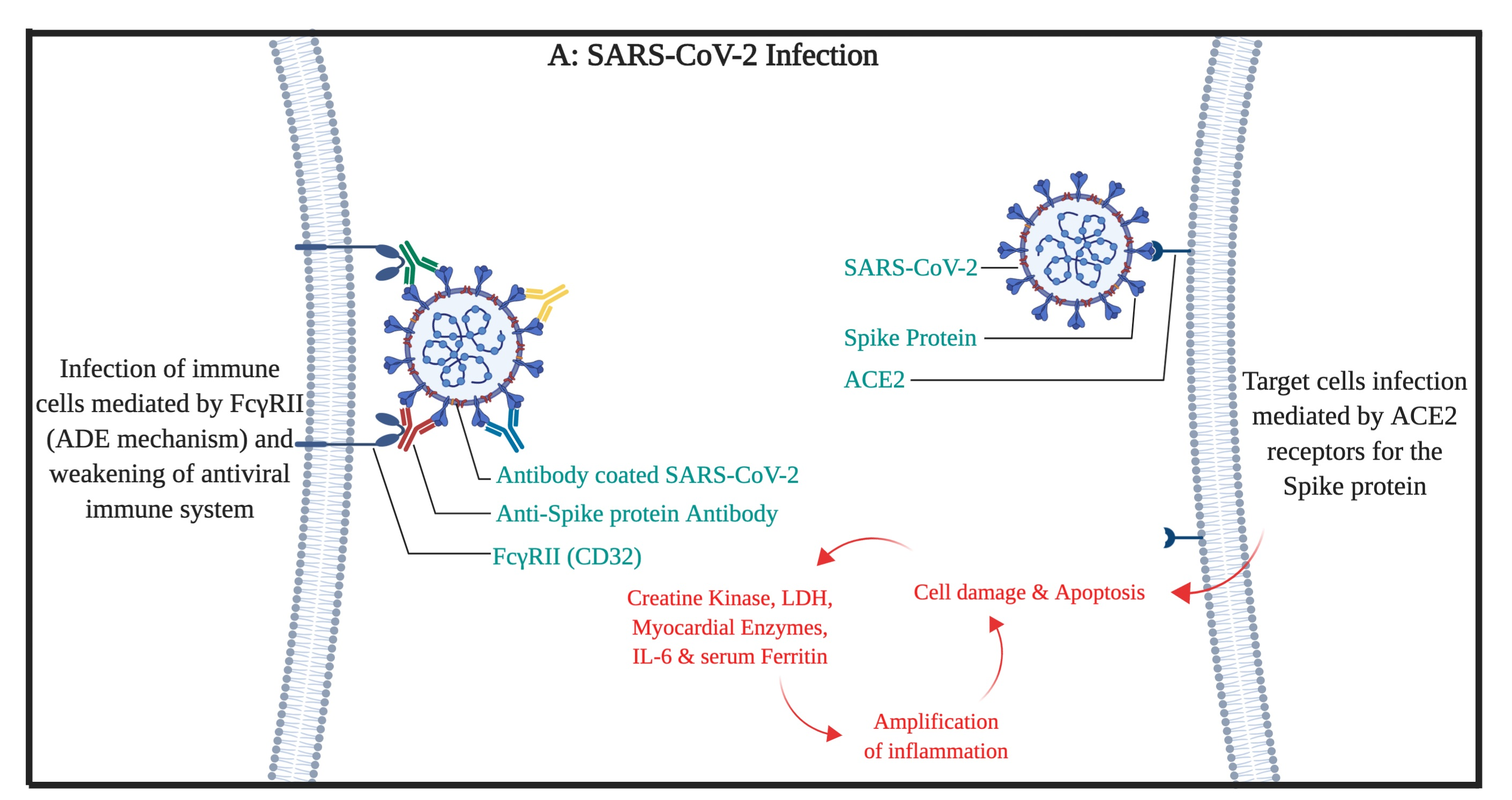
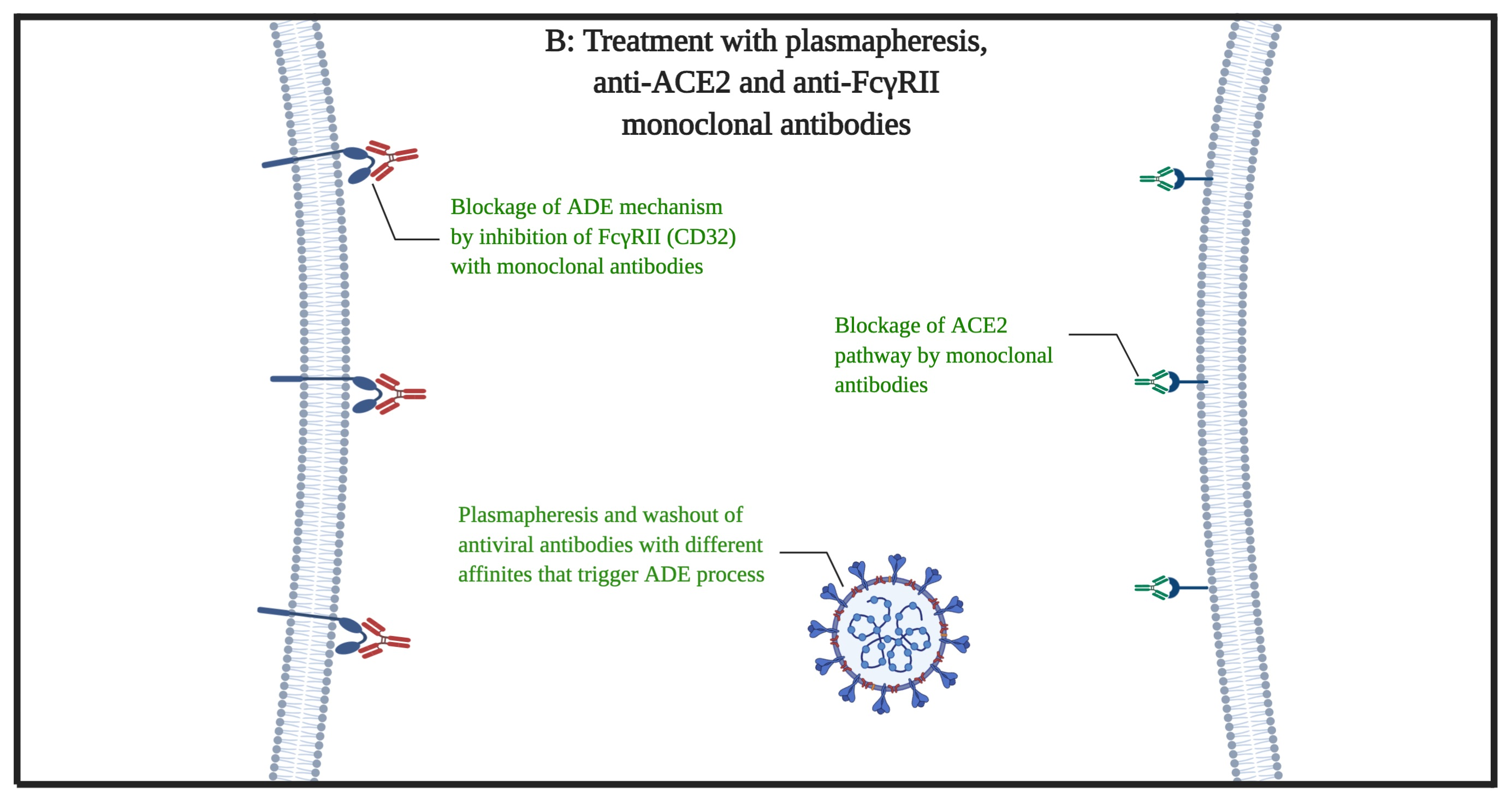
In March 2020, the WHO declared the COVID-19 disease as pandemic disease. There have been studies on the COVID-19 to find a certain treatment but yet, there is no certain cure. In this article, we present a possible way to treat severe cases of COVID-19. Based on previous studies, there are similarities between the Spike antigens of SARS-CoV and SARS-CoV-2 viruses. It is expected that these similarities (structural and affinity to the receptor of ACE2) can lead to the same pathophysiological activity of the virus by the use of ACE2 and FcγRII (the antibody-dependent enhancement mechanism). Therefore, we propose a way of washing out (by plasmapheresis) the possible antibodies against the spike protein of the virus out of patients plasma to stop the antibody-dependent enhancement (ADE) mediated infection of the immune system cells at the first phase of the treatment and simultaneous use of the anti-ACE2 with anti-FcγRII monoclonal antibodies at the second phase. We propose these procedures for the patients that has no significant response for typical anti-viral, ARDS, and conservative therapies and the disease persists or progresses despite sufficient therapies.
Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2
Gaurav Sahay, PhD, Oregon Stare Univ
Oregon Stare Univ
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters through the airways and infects the lungs, causing lethal pulmonary damage in vulnerable patients. This virus contains spike proteins on its envelope that binds to human angiotensin-converting enzyme 2 (hACE2) expressed on the surface of airway cells, enabling entry of the virus for causing infection. In severe cases, the virus enters the circulatory system, contributing to multiorgan failure. Soluble form of ACE2 binds to SARS-CoV-2 spike protein and prevents viral entry into target cells. Moreover, recombinant soluble ACE2 ameliorates lung injury but its short half-life limits its therapeutic utility. Here, we engineered synthetic mRNA to encode a soluble form of hACE2 (hsACE2). to prevent viral infection. Novel lipid nanoparticles (LNPs) were used to package mRNA and transfect mammalian cells for enhanced production of secreted proteins. A single dose of intravenously administered LNP led to hepatic delivery of the mRNA. This elicited secretion of hsACE2 into the blood circulation within 2 h, and levels of circulating hsACE2 peaked at 6 h and gradually decreased over several days. Since the primary site of entry and pathogenesis for the SARS-CoV-2 is the lungs, we instilled LNPs into the lungs and were able to detect hsACE2 in the bronchoalveolar lavage fluid within 24 h and lasted for 48 h. Through co-immunoprecipitation, we found that mRNA-generated hsACE2 was able to bind with the receptor binding domain of the SARS-CoV-2 spike protein. Furthermore, hsACE2 was able to strongly inhibit (over 90%) SARS-CoV-2 pseudovirus infection. Our proof of principle study shows that mRNA-based nanotherapeutics can be potentially deployed for pulmonary and extrapulmonary neutralization of SARS-CoV-2 and can open new treatment opportunities for COVID-19.
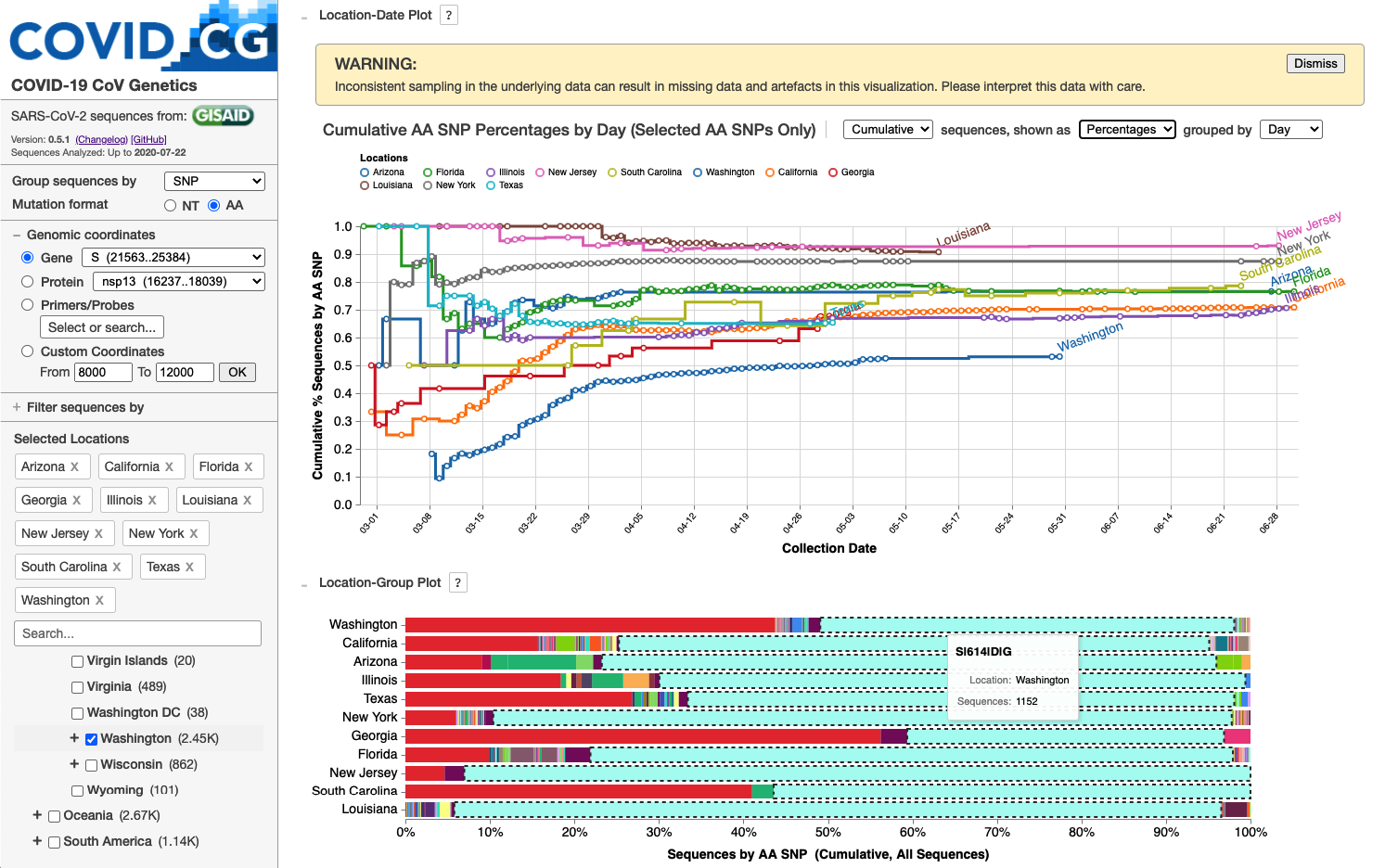
COVID-19 CG: Tracking SARS-CoV-2 by Mutation, Location, and Date of Interest
Albert Chen, Broad Institute of MIT and Harvard1, Shing Zhan, University of British Columbia2, Benjamin Deverman, PhD, Broad Institute3, Alina Chan, PhD, Broad Institute3, Kevin Altschuler
Broad Institute of MIT and Harvard1
University of British Columbia2
Broad Institute3
We developed the COVID-19 CoV Genetics (COVID-19 CG at covidcg.org) browser to enable users to track SARS-CoV-2 single-nucleotide variations (SNVs) and lineages while rapidly filtering by location, date, and mutation of interest. COVID-19 CG aims to provide significant time, labor, and cost-saving utility to the diverse projects on SARS-CoV-2 transmission, evolution, emergence, immune interactions, diagnostics, therapeutics, vaccines, and tracking of interventions. Via serverless deployment, users can view the comprehensive nucleotide and amino acid residue variation in their selection to inform their research or product development. No existing public browsers or software provide these capabilities. To accelerate COVID-19 research and public health efforts, COVID-19 CG will be continually upgraded with new and improved features so that users can quickly and reliably pinpoint critical mutations as the virus evolves throughout the pandemic. Towards this goal, we strongly advocate that countries continue or increase their sequencing of SARS-CoV-2 isolates from their patients (and infected animals) and share this data in a timely manner so that scientists worldwide are maximally informed about developments in the spread of SARS-CoV-2.

Approved drugs inhibiting TMEM16 proteins block SARS-CoV-2 Spike-induced cellular syncytia
Mauro Giacca, MD, PhD, King's College London1, Luca Braga, , Hashim ALI, KCL2
King's College London1
KCL2
COVID-19 is a disease with unique characteristics. In addition to the clinical features common to interstitial pneumonias and other causes of acute respiratory distress syndrome (ARDS), this condition is characterized by lung thrombosis, frequent diarrhoea, and a set of unexplained symptoms, such as unperceived low oxygen saturation (happy hypoxia), loss of smell and taste and rapid deterioration of lung function consistent with alveolar oedema. These features suggest that the disease has a more complex explanation than simply death of pneumocytes caused by SARS-CoV-2 replication. While some of these findings can be attributed to abnormal activation of the inflammatory or immune response, the pathological substrate for this response still remains largely elusive.
First, we investigated 41 consecutive post-mortem samples from individuals who died of COVID-19. We found that lung pathology is characterized by extensive alveolar damage and thrombosis of the lung micro- and macro-vasculature. Pneumocytes and endothelial cells contained viral RNA even at the later stages of the disease. An additional, common feature was the common presence of a large number of dysmorphic pneumocytes, often forming syncytial elements. Generation of these syncytia likely results from the expression and activation of the SARS-CoV-2 Spike protein on the infected cell plasma membrane.
Based on these observations, we performed two high-content microscopy-based, high-throughput screenings with over ~3000 approved drugs to search for inhibitors of Spike-driven syncytia. We converged on the identification of 83 drugs that inhibited Spike-mediated cell-cell fusion, several of which belonged to defined pharmacological classes. Of these, we focussed our attention on effective drugs that also protected the cells against virus-induced death. In particular, we found that Niclosamide, the top drug in our screenings, markedly blunted calcium oscillations and chloride channel currents in Spike-expressing cells by blocking the activation of members of the TMEM16/Anoctamin family of proteins. Consistent with this observation, downregulation of the ion channel and scramblase TMEM16F blocked Spike-induced syncytia formation. Together, these findings support the possibility that COVID-19 disease is due to the prolonged persistence of virus-infected cells, suggest potential mechanisms for COVID-19 disease pathogenesis and sustain the repurposing of Niclosamide for therapy.
Developing Oligonucleotide Therapeutics for Targeted Delivery and Silencing of Conserved SARS COV-2 Gene Regions
Vignesh Narayan Hariharan, PhD, University of Massachusetts Medical Scho1, Minwook Shin, , Pranathi Krishnamurthy, , Annabelle Biscans, , Daniel O'Reilly, , Qi Tang, , Kathryn Monopoli, , Chantal Ferguson, , John Cruz, , Mohan Somasundaran, , Jill Perreira, , Gitali Devi, , William Mcdougall, , Sarah Davis, , Samuel Hildebrand, , Bruno Godinho, , Robert Finberg, , Jennifer Wang, , Jonathan Watts, , Anastasia Khvorova, PhD, University of Massachusetts Medical Scho1
University of Massachusetts Medical Scho1
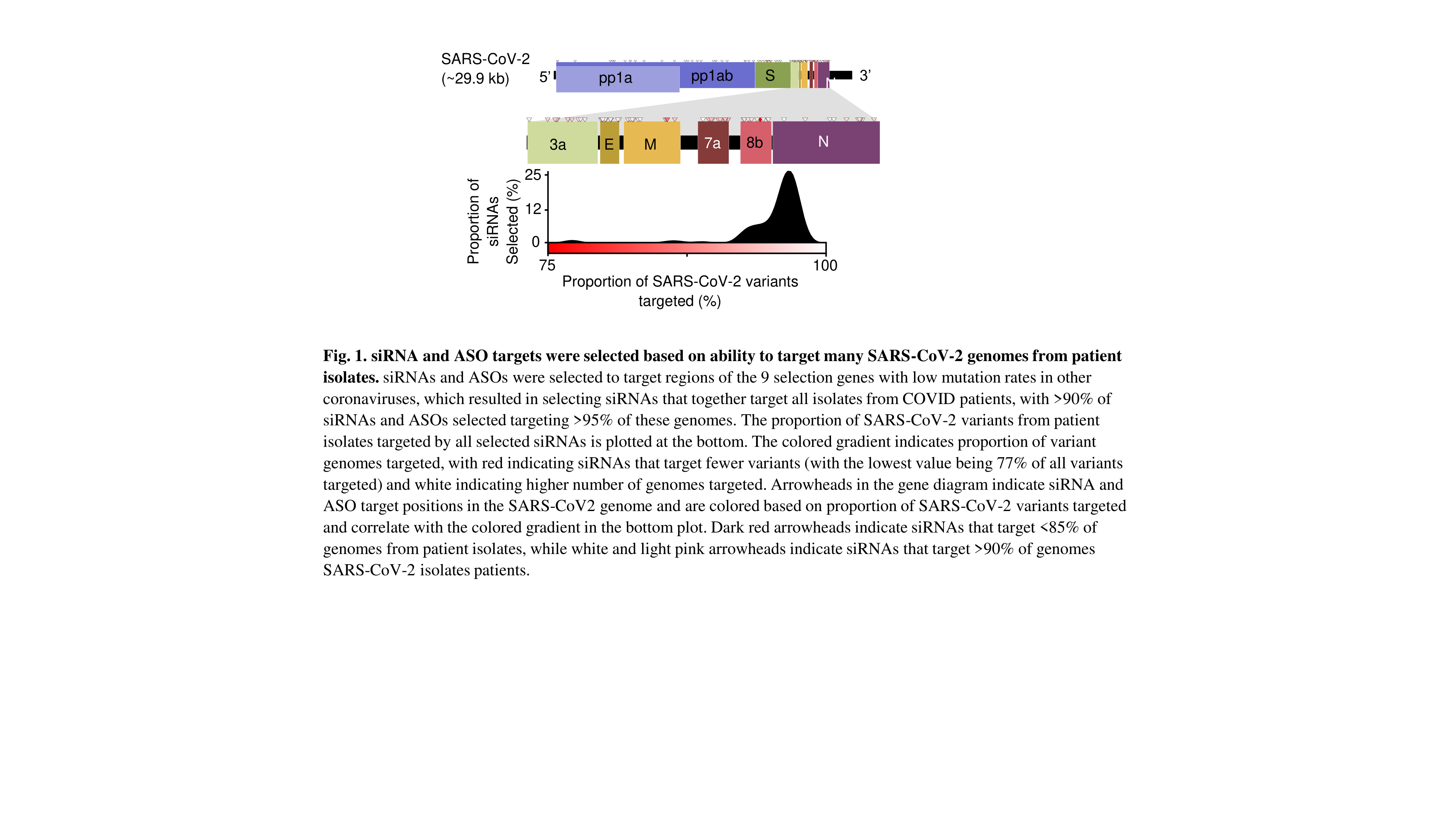
The rapid spread of SARS CoV-2 throughout the globe has resulted in over 17 million infections and 600000 deaths worldwide with no signs of abatement. The continued rise of infection and mortality rates has crippled healthcare and devastated economic systems, with a slow and long road to recovery ahead. At present, there is no vaccine against infection and therapeutic avenues are limited. Oligonucleotide therapeutics, specifically small interfering RNA (siRNA) and antisense oligonucleotides (ASO) are uniquely poised to combat the spread of SARS COV-2 since they can be rapidly and rationally designed to re-direct endogenous pathways (RNA interference or RNase H) to degrade specific viral targets. Additionally, both ASOs and siRNAs can be delivered to specific tissues, including the lung, using conjugation and chemical engineering.
Here, we describe the development and optimization of oligonucleotides targeting multiple regions of the SARS-CoV-2 mRNA to achieve protection against viral infection in vitro. After screening of hundreds of variants, siRNAs and ASOs have been developed against conserved genetic regions, allowing at least 95% of 718 SARS-CoV-2 patient isolates (Fig. 1). Lead siRNA and ASO compounds achieve greater than 99% reduction in viral RNA and at least 90% reduction in viral proteins in a VERO E6 model of infection. Furthermore, we show that chemical engineering of siRNA and ASO scaffolds enables sustained lung accumulation (Fig. 2) and target gene silencing after systemic (subcutaneous) or local (intratracheal) administration.
Taken together, we have developed an oligonucleotide chemistry platform and identified lead candidates with potential to protect (when administered as a prophylactic) or limit infection with SARS-CoV-2. Preliminary experiments in SARS-CoV-2-infected non-human primates are underway.

A Single-shot anti-SARS-CoV-2 Vaccine Based on a Novel Gorilla-Adenoviral Vector Stabilized in a Tailored Liquid Formulation
Alessandra Vitelli, , Eva Reinauer, , Julia Rabas, , Sabine Hauck, PhD, Leukocare1, Stefano Colloca
Leukocare1
As a new vaccine against COVID-19, a novel simian adenoviral vector was developed. This vector is based on a proprietary vaccine platform technology based on species C simian adenoviral (sAd) vectors derived from chimpanzee, bonobo and more recently from gorilla (GRAd). Species C simian Ad vectors have been developed as vaccine candidates for multiple infectious diseases and there is ample preclinical and clinical evidence that these vectors are safe and induce antigen-specific cellular and humoral immunity in all age groups. Importantly, sAd vectors belonging to species C adenovirus and thus related to human Ad5 (hAd5) demonstrated higher immunological potency and protective efficacy in animal model in comparison to vector systems derived from other Ad species.
Differently from the highly potent hAd5, sAds are not hampered by the pre-existing immunity against hAd vectors. In addition to inducing strong antibody responses, sAd-vector encoded antigens are known to elicit vigorous cellular responses, which can help to strengthen the adaptive immunity and to sustain the humoral responses. This makes them particularly suitable for a single dose approach to rapidly induce protective immunity during an outbreak, as was shown by the protective efficacy of a single intramuscular (IM) injection with the chimpanzee-derived adenoviral vector ChAd3 encoding Ebola virus glycoprotein (EBOV GP) against acute lethal challenge in NHP: When evaluated in clinical trials, ChAd3 Ebola vaccine induced neutralizing antibody titres comparable to those induced by the protective VSV Ebola vaccine, besides a better safety profile in humans (1).
Specifically for the new vaccine against COVID-19, the recently developed novel proprietary gorilla adenovirus (GRAd32) will be employed that demonstrated high immunogenicity in mice and that is poorly cross-neutralized by human sera with respect to hAd5 or to other species C adenovirus. This replication-defective GRAd was transfected with a transgene encoding for the pre-fusion stabilized SARS-CoV2 spike glycoprotein, which is the viral protein responsible for the attachment of the virus to the host cellular receptor and the main target of neutralizing antibodies. This virus is currently formulated in a buffer for frozen storage.
For improved storage conditions, a stabilizing formulation should be developed based on formulations tailored for a hAd5 in a DOE-based development approach (2). The formulations identified in this approach were investigated for their stabilizing potential on the GRAd. The experimental results from accelerated aging studies showed comparable stabilizing properties of these human Ad5 formulations also for the simian adenoviral vector. However, the stabilizing potential might be dependent on the actual transgene and, therefore, a tailored formulation is considered beneficial for the viral vector-based vaccine for best stabilization. Especially in times of manufacturing capacity bottlenecks an extended storage time and little limitations in transport conditions are important to ensure vaccine availability.
New prognostic algorithms for differential diagnostics in SARS‑CoV‑2 infection
Ekaterina Semina, PhD, Cardiology Research Cdenter1, Nailya Sabitova, Lomonosov Moscow state University2, Natalya Borovkova, , Yuli Andreev, , Aslan Shabanov, , Kseniya Rubina, DSci,PhD, Lomonosov Moscow State University2
Cardiology Research Cdenter1
Lomonosov Moscow state University2
The pandemic situation with COVID-19 - new coronavirus infection due to SARS-CoV-2, requires an integrated approach, including the creation of vaccines to prevent the spread of the disease, the development of new drugs aimed at lowering the viral load, suppressing of an excessive immune response, preventing the pulmonary fibrosis and the disseminated intravascular coagulation. The development of new prognostic algorithms for differential diagnostics to predict and reduce the severity and complications of COVID-19 remains extremely relevant.
It should be noted that the severe course of the disease has been registered not only in elderly patients or patients suffering from comorbidities but also in healthy young people. One of the promising targets for studying COVID-19 complications such as pneumonia, systemic inflammation and disseminated intravascular coagulation is the hemostasis system. We conducted a comprehensive study using blood serum from patients (N = 60) with confirmed COVID-19 admitted to N.V. Sklifosovsky Research Institute for Emergency Medicine, Moscow, within the last 4 months (from April to July 2020). The levels of plasminogen (Plg), plasminogen activator inhibitor 1 PAI, IL-1a, -17, TGFb, TNFa, and adiponectin in blood serum were correlated with apoptotic markers in blood leukocytes (AKT, BAD, BCL-2, CASPASE-8, -9, JNK and P53), with CT lung scans, BMI, duration of mechanical ventilation and mortality. Our preliminary data show a consistent relationship between apoptosis of blood cells, BMI and mortality.
Important Role of Gamma-Delta T Cells in Anti-SARS-CoV-2 Immune Response
Serhat Gumrukcu, MD, PhD, Seraph Research Institute1, Spriha Singh, , Gregory Howell, BA, Seraph Research Institute1, Phillip Musikanth, MD, Seraph Research Institute1, Tung Nguyen
Seraph Research Institute1
Background
COVID-19 is the most devastating pandemic in this century. Inconsistent and short-lived antibody responses are concerning. It has been shown that gamma-delta (gd) T cells played a significant role in past SARS epidemics. We hypothesized that cellular immunity could also play a major role in SARS-CoV-2 infection. Therefore, we evaluated the SARS-CoV-2-specific antiviral activities of gdT cells isolated from COVID-19 convalescent individuals.
Methods
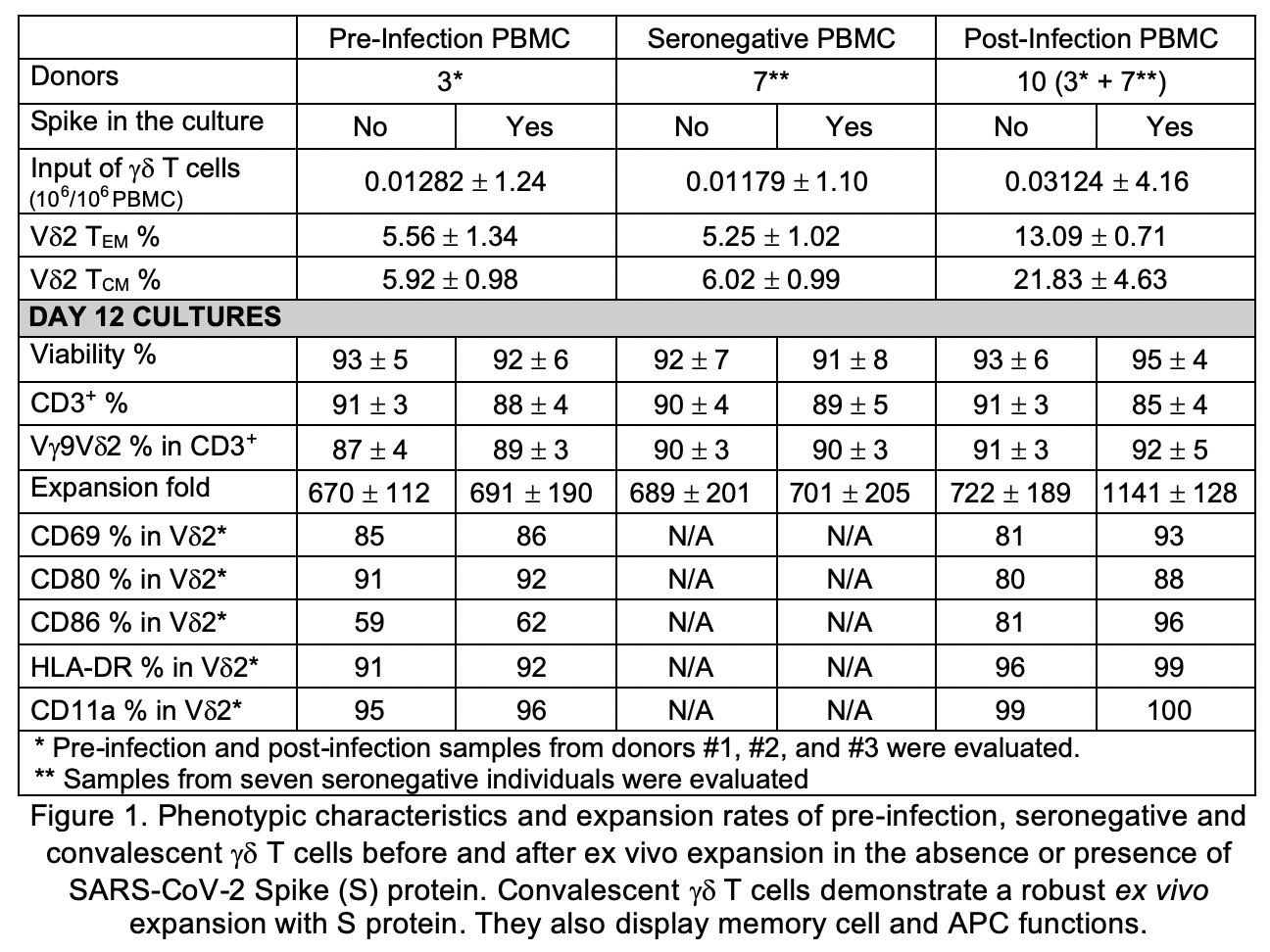
We analyzed plasma and PBMC from ten trial participants with confirmed COVID-19 infection within the past 90 days and seven seronegative individuals. Cell-mediated immune responses were evaluated by screening all PBMC subsets by flow cytometry, including alpha-beta (ab) and gd T-cell receptor (TCR) repertoires using 24 TCR Vb and 3 TCR Vd chain-specific monoclonal antibodies (MAbs). Pre-infection PBMCs from 3 out of 10 trial subjects were also compared to their respective post-infection samples by immunophenotyping via flow cytometry. Pre- and post-infection gd T cells were expanded ex vivo in the presence or absence of 100nM recombinant SARS-CoV-2 spike (S) protein and evaluated for expansion rate, purity and CD45RA-CD27-effector memory (EM) and CD45RA-CD27+ central memory (CM) subset percentages. On day 3 of expansion, supernatants were analyzed by a flow-based multiplex assay to quantify IL-2, IL-4, IL-10, IL-6, IL-17A, TNF-α, sFas, sFasL, IFN-γ, granzyme A, granzyme B, perforin and granulysin.
Results
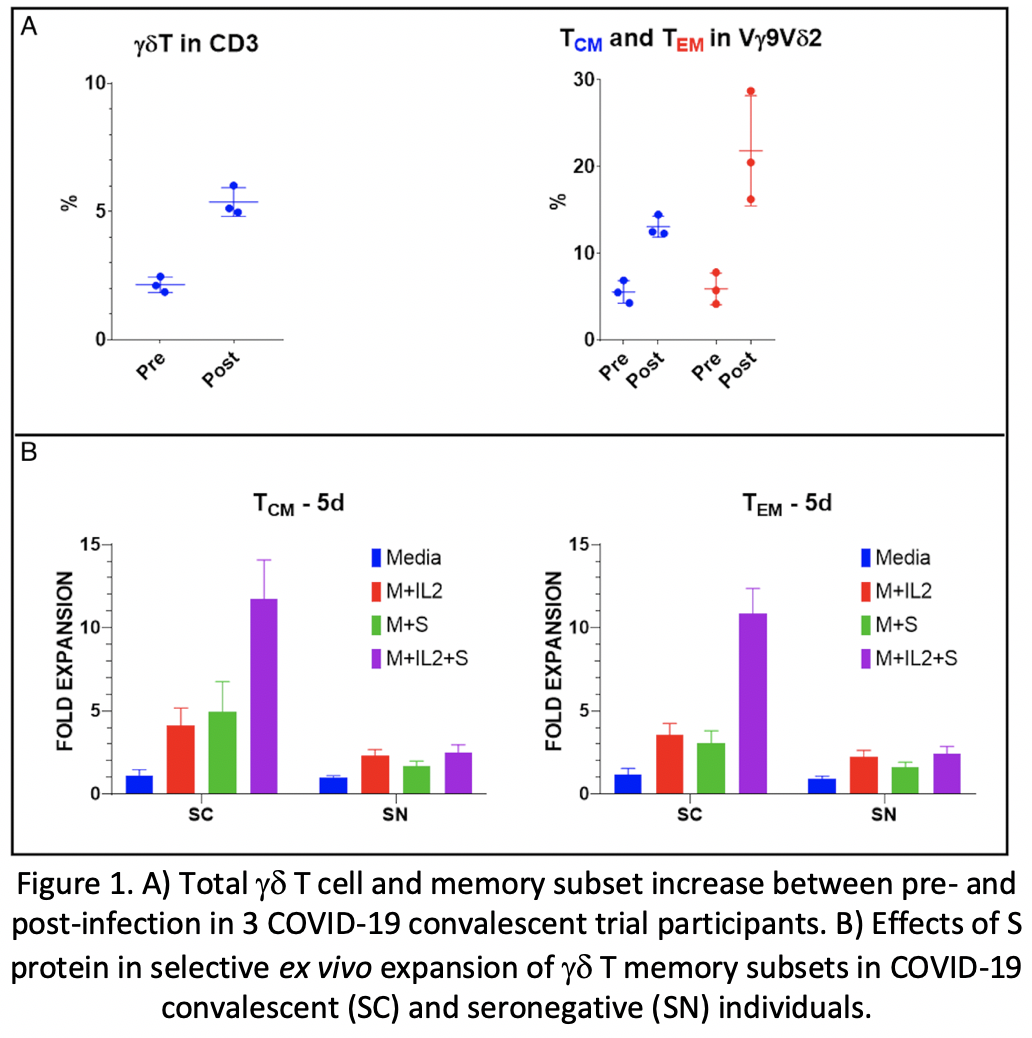
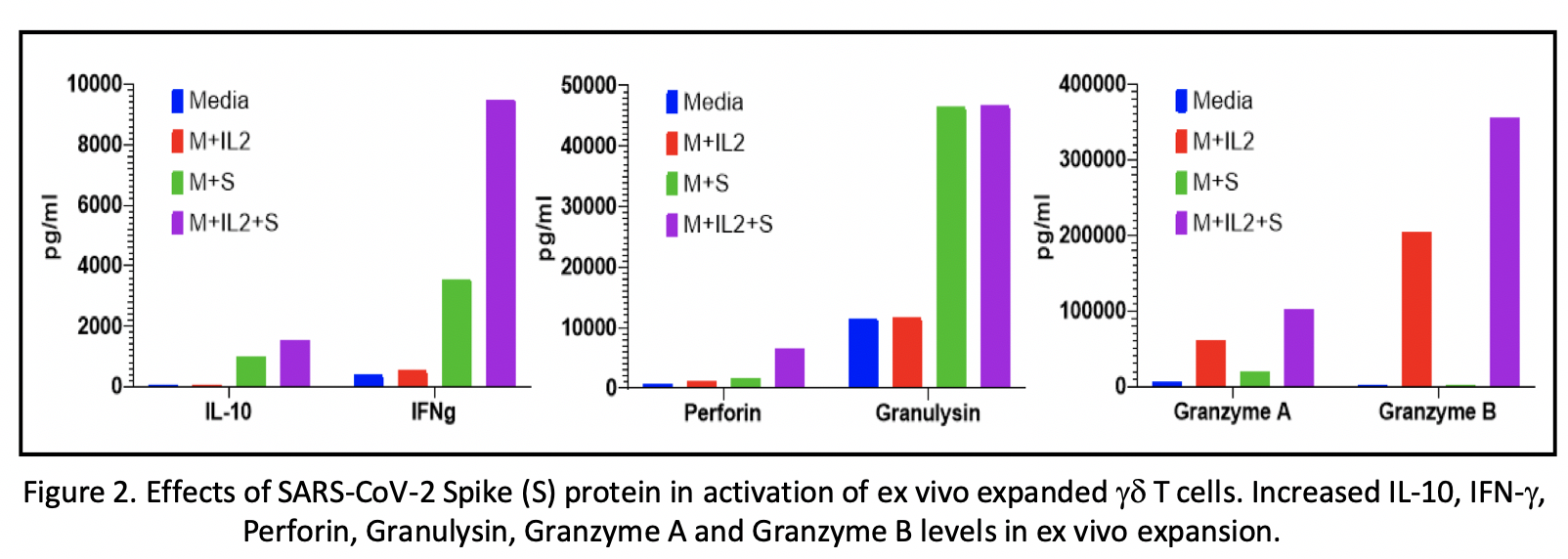
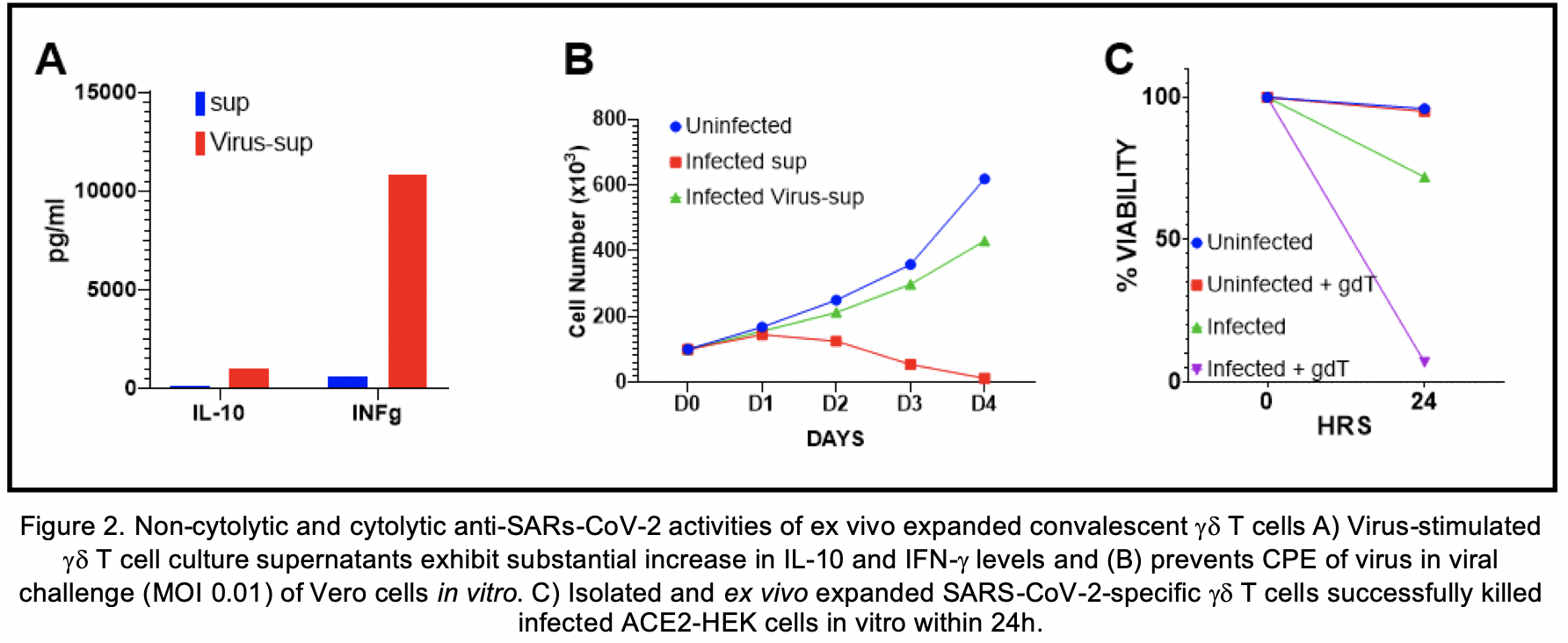
Analyses of the immunophenotyping and T cell repertoires in all ten convalescent patients revealed that gd T cell populations were activated during the infection. The comparison between pre- and post-infection immune cell profiles in several subjects demonstrated that certain memory subsets of Vg9Vd2 T cell populations were also selectively expanded (250%, 975%, and 617% absolute count increase in gd T, Vd2 EM, and Vd2 CM cells respectively) (Fig 1A). There was no change in TCR Vb chain repertoire post-infection. Ex vivo expansion of pre- and post-infection gd T cells in the presence or absence of S protein revealed substantial expansion of EM and CM gdT cells in the post-infection PBMC cultures supplemented with S protein compared to other groups (Fig 1B). Multiplex cytokine analyses showed increases in IFN-g, IL-10, granzyme A, granzyme B, perforin and granulysin in gd T cell cultures supplemented with S protein compared to other groups (Fig 2).
Conclusion
Substantial increases in gd T cell populations were observed in post-COVID19 PBMCs compared to pre-COVID19 and seronegative samples. These cells expanded robustly in the presence of S protein and demonstrated memory cell properties, suggesting an important role of gd T cells in sustained immunity against SARS-CoV-2. Our findings, supported by similar reports in past SARS epidemics, suggest potential therapeutic and preventative applications of SARS-CoV-2-specific gd T cells. Therapeutic and/or prophylactic cell-based vaccine approaches are under development.
A Serotypically Evolved Recombinant Measles Virus as a COVID-19 VaccineCandidate for High Risk/Older Individuals
Miguel Muñoz-Alía, PhD, Mayo Clinic/Department of Molecular Medicine1, Rebecca Nace, , Nandakumar Packiriswamy, PhD, Vyriad2, Baskar Balakrishnan, PhD, , Stephen Russell, MD, PhD, Mayo Clinic3
Mayo Clinic/Department of Molecular Medicine1
Vyriad2
Mayo Clinic3
A successful vaccine against the severe acute respiratory syndrome coronavirus (SARS-CoV-2) would provide an effective containment for the current massive pandemic. The elderly are among the most vulnerable to the coronavirus disease 2019 (COVID-19) and a highly immunogenic and effective vaccine in this group population is urgently needed. Here, we described a human-serum neutralization resistant measles virus (MeV) expressing various SARS-CoV-2 spike (S) protein antigens as self-assembling nanoparticles. Additionally, we studied the immunogenicity of the different SARS-CoV-2-S subdomains by immunizing IFNAR-/--CD46Ge mice with the various antigens: S ectodomain (S1+S2, amino acids 16 to 1213), S1 subdomain (amino acids 16-213), S1-receptor binding domain (RBD, amino acids 319-541) and S2 subdomain (amino acids 686-1213). All these antigens elicited fusion-inhibiting antibodies, with the RBD immunogen trending towards higher potency. Paradoxically, antibodies generated by the RBD could not be detected by enzyme-linked immunosorbent assay (ELISA). The same analysis showed that the S1+S2 ectodomain and S2 generated 28-fold higher antibody titers than those generated by the S1 subdomain. In the context of the MeV vector, the introduction of prefusion stabilizing mutations into the S1+S2 ectodomains and genetic fusion to ferritin increased 2-fold the protein expression while maintaining unchanged the growth kinetic of the virus. Mice immunization with this MeV-based prefusion SARS-CoV-2 S1+S2 nanoparticle vector resulted in the highest IgG antibody response. Thus, a stealthy MeV displaying SARS-CoV-2 S as self-assembling nanoparticles might be a feasible option for immunization of the elderly with pre-existent immunity against the vector.
Therapeutic and Protective Immunotherapy Potential of SARS-CoV-2-Specific Gamma-Delta T Cells
Serhat Gumrukcu, MD, PhD, Seraph Research Institute1, Gregory Howell, BA, Seraph Research Institute1, Phillip Musikanth, MD, Seraph Research Institute1, Tung Nguyen
Seraph Research Institute1
Background
COVID-19 is the most devastating pandemic in this century. Inconsistent and short-lived antibody responses are concerning. Gamma-delta (gd) T cells were shown to have an important role in anti-SARS-CoV immunity in past SARS epidemics. We have hypothesized and recently demonstrated their significance in anti-SARS-CoV-2 immunity. In this study, we evaluated their potential as therapeutic and protective cell-based immunotherapy.
Methods
gd T cells were expanded ex vivo from pre- and post-infection PBMCs in the presence or absence of 100nM recombinant SARS-CoV-2 spike (S) protein. To assess their non-cytolytic antiviral activities, expanded cells were exposed to SARS-CoV-2-infected Vero cell culture supernatants. On day 4 after the exposure, gd T cell supernatants were added to Vero cell cultures 24h before infecting them with SARS-CoV-2 (MOI 0.01). Cytopathic effects (CPE) of the virus were measured by daily cell count and imaging. Expanded gd T cells were co-cultured 1:1 with infected (MOI 1) and uninfected ACE2-HEK cells to test their cytolytic antiviral activity. After 24h, target cell viability was measured by cell counter and flow cytometry. Co-culture supernatants were analyzed by a flow-based multiplex cytokine assay. Expanded gd T cells were also assessed for markers for activation by CD25 and CD69, for cell exhaustion by CD57, and for antigen presenting cell (APC) functions by CD80, CD86, HLA-DR, CD11a, CXCR5, and CCR7 with flow cytometry. CD45RA-CD27- effector memory (EM) and CD45RA-CD27+ central memory (CM) subsets of expanded gd T cells were enriched by immunomagnetic separation and co-cultured with B cells harvested from seronegative individuals for assessment of their vaccine effects in vitro. Antibodies in co-culture supernatants were measured and evaluated for neutralization activity.
Results
Expansion rate of gd T cells, and their EM and CM memory subsets were substantially higher in post-infection PBMCs compared to pre-infection and seronegative controls. Vg9Vd2 T cell populations were selectively expanded in ex vivo cultures supplemented with S protein compared to other groups. Moreover, memory subsets of ex vivo expanded SARS-CoV-2-specific gd T cells displayed potent APC functions, suggesting not only therapeutic, but also potential protective immunotherapy implications of these cells (Fig 1). Virus-stimulated gd T cell culture supernatants exhibited increased IFN-g, IL-10 levels and prevented CPE of virus at in vitro viral challenge (Fig 2A-B). Isolated expanded gd T cells effectively killed infected cells in vitro (Fig 2C). In vitro vaccinated B cells demonstrated a robust production of SARS-CoV-2 neutralizing antibodies, predominantly in IgM and IgA isotypes. Detailed data will be presented.
Conclusions
SARS-CoV-2-specific gd T cells harvested from convalescent individuals exhibited strong non-cytolytic and cytolytic antiviral activities, as well as substantial ex vivo expansion capabilities. Moreover, memory subsets of these cells displayed enhanced APC functions and elicited robust neutralizing antibody production in B cells harvested from seronegative individuals. Our findings support potential clinical use of these cells for treatment and prophylaxis of COVID-19. Our cell-based immunotherapy and vaccine candidates generated from SARS-CoV-2-specific convalescent gd T cells are currently undergoing development and in vivo testing. We are also exploring larger scale approaches based on our findings.
Establishment of human airway organoids for COVID-19 drug and vaccine development
Kazuo Takayama, Dr, Kyoto University1, Toru Okamoto
Kyoto University1
Coronavirus disease 2019 (COVID-19) is a disease that causes fatal disorders including severe pneumonia. To develop a therapeutic drug for COVID-19, a model that can reproduce the viral life cycle and evaluate the drug efficacy of anti-viral drugs is essential. In this study, we established a method to generate human bronchial organoids (hBO) from commercially available cryopreserved human bronchial epithelial cells and examined whether they could be used as a model for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) research. Our human bronchial organoids contain basal, club, ciliated, and goblet cells. Angiotensin-converting enzyme 2 (ACE2), which is a receptor for SARS-CoV-2, and transmembrane serine proteinase 2 (TMPRSS2), which is an essential serine protease for priming spike (S) protein of SARS-CoV-2, were highly expressed in human bronchial organoids. After SARS-CoV-2 infection, not only the intracellular viral genome, but also progeny virus, cytotoxicity, pyknotic cells, and moderate increase of the type I interferon signal could be observed. Treatment with camostat, an inhibitor of TMPRSS2, reduced the intracellular viral copy number to 2% of the control group. We are currently evaluating other drugs in human bronchial organoids. Furthermore, the gene expression profile in SARS-CoV-2-non-infected or -infected human bronchial organoids was obtained by performing RNA-seq analysis. In conclusion, we succeeded in generating human bronchial organoids that would be used for SARS-CoV-2 research and COVID-19 drug and vaccine development.
Evaluation of Losartanids Administration on SARS-Cov-2 Receptor (ACE2) Expression and Possible Therapeutic Inhibition for the Reduction of Virus Transmission
FELICE AMATO, PhD, CEINGE BIOTECNOLOGIE AVANZATE1, Marika Comegna, , Maria Vitale, University Federco II of Naples2, Immacolata Zollo, , Giuseppe Castaldo, , Lucio Pastore
CEINGE BIOTECNOLOGIE AVANZATE1
University Federco II of Naples2
The COVID-19 pandemic caused by the new coronavirus SARS-CoV-2 has killed thousands of people worldwide. Currently, no specific treatments are available, and therapies being used are entirely supportive and reliant on patient health and history. In humans, CoVs tend to cause mild to moderate upper respiratory tract infections such as the common cold. These strains exhibited stronger virulence and quickly passed from human to human. The entry receptor utilized by SARS-CoV-2 is Angiotensin-Converting Enzyme-2 (ACE-2). ACE-2 is a type I transmembrane metallocarboxypeptidase with homology to ACE, an enzyme long-known to be a key player in the Renin-Angiotensin system (RAS) and a target for the treatment of hypertension. ACE-2 is mainly expressed in vascular endothelial cells, the renal tubular epithelium, and in Leydig cells in the testis and also in the lung, kidney, and gastrointestinal tract. ACE-2 has also been shown to exhibit a protective function in the cardiovascular system and other organs. ACE and angiotensin receptor blockers (ACE inhibitors or ARBs), drugs normally used to treat pathologies such as hypertension, diabetes, and cardiovascular diseases, cause an upregulation of ACE-2 and it is still not yet clear if they have the potential to worsen the infection. However, this could explain why COVID-19 patients with these pathologies have the highest mortality rate.
To evaluate the effect of Losartan, an ACE-2 antagonist, on the expression of ACE-2 mRNA, we treated lung epithelial cells and nasal mucosa cells using different concentrations of this yet commercially available drug (100 nM, 300 nM and 900 nM). The data showed a slight increase in ACE-2 expression levels in polarized nasal mucosa cells.
In addition, we propose a novel approach to reduce the spreading of SARS-CoV-2 infection based on silencing its receptor, ACE-2, in nasal and oro-faringeal epithelium using short interfering RNAs (siRNAs). We treated lung epithelial cells and nasal mucosa cells with ACE-2 siRNA combination and verified the reduction of ACE-2 expression by RT-PCR. Combining the siRNAs with chitosan into nanoparticles with high efficiency, we plan to produce a pharmaceutical formulation, such as a nasal spray, that could be clinically evaluated as soon as possible; in fact, both chitosan and siRNA can be produced and therefore purchased at clinical grade quality. Local delivery of ACE-2 inhibitor should be able to reduce viral receptor expression, thereby reducing susceptibility to the virus, without alterations of blood pressure or other systemic side-effects.
1.Lullo, A. Di et al. An “ ex vivo model ” contributing to the diagnosis and evaluation of new drugs
in cystic fibrosis. Acta Otorhinolaryngol. Ital. 1–7 (2016). doi:10.14639/0392-100X-1328
2. Napolitano M. et al. Comparative Analysis of Gene Expression Data Reveals Novel Targets of Senescence-Associated microRNAs. PLoS One. 2014 Jun 6;9(6) doi: 10.1371/journal.pone.0098669.
Mannose mimicking lipid nanoparticle encapsulated spike mRNA as a vaccine for SARS-CoV-2 virus
Srujan Kumar Marepally, PhD, Center for Stem Cell Research, CMC-Vellore1, Porkizhi Arjunan, MSc, , Ajay Kumar Dhyani, MSc, , Aruna Mohan, MSc, , Mohan Kumar Murugesan Kumarasamypet, PhD, , George Varghese, , Alok Srivastava, MD, Christian Medical College2
Center for Stem Cell Research, CMC-Vellore1
Christian Medical College2
Rapid and large-scale deployment of vaccines is challenging with conventional vaccines for pandemics such as SARS-COV-2. Antigen encoding mRNA vaccines offer great advantage as they are non-immunogenic, non-integrative and less expensive. mRNA based vaccines developed in shorter time and found be effective in the recent SARS-COV-2 outbreak, and are the first to get approvals for clinical testing. Moderna therapeutics leading from the front followed by BioNTech, CurVac and Arcturus therapeutics are using lipid nanoparticles encapsulated spike mRNA as potential vaccine candidates for SARS-CoV2. In lines with the current approaches, we are developing a potential vaccine candidate with our customized lipid nanoparticle system and UTR optimized, chemically modified spike mRNA (Figure 1). In our prior findings, we demonstrated that tethering mannose mimicking ligands to lipid nanoparticle system imparts specificity to dendritic cell targeting delivery through mannose receptor mediated endocytosis and trigger immune responses. Taking cues from our prior findings, we developed novel shikimoylated mannose receptor targeting nanoparticle system (SMART nanoparticles) that could effectively deliver nucleic acids in DCs (Figure 2A). We also identified 5’ and 3’ UTRs that are as efficient as Moderna therapeutics reported UTRs (Figure 2B). Later, we synthesized chemically modified mRNA with encoding spike protein, a surface antigen of SARS-CoV-2 and evaluated their expression in vitro with ours lipid nanoparticle system (Figure 2C). We will also discuss our on-going mRNA vaccine development with DC targeted lipid nanoparticle system.
A COVID-19 intradermal (ID) delivered DNA vaccine is immunogenic in multiple animal models and provides anamnestic protection in rhesus macaques against SARS-CoV-2 infection
Ami Patel, PhD, The Wistar Institute1, Jewell Walters, , Emma Reuschel, PhD, , Katherine Schultheis, , Elizabeth Parzych, PhD, , Ebony Gary, PhD, The Wistar Institute1, Stephanie Ramos, Inovio Pharmaceuticals2, Trevor Smith, PhD, , David Weiner, PhD, The Wistar Institute1, Kate Broderick
The Wistar Institute1
Inovio Pharmaceuticals2
The coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2 coronavirus, has had a dramatic impact on global health, with tremendous repercussions to social and economic infrastructures. COVID-19 disease presents as a mild-to-moderate respiratory illness that resolves in >80% of people, however severe disease progression in the remaining 20% is associated with a hyperinflammatory response, acute respiratory distress, potentially leading to systemic organ failure and death. There is therefore an urgent need for preventative vaccines and treatments to reduce the impact of COVID-19 disease. We developed a synthetic DNA vaccine (INO-4800) targeting the SARS-CoV-2 Spike protein that induces robust immune responses in mice, guinea pigs, rabbits, and rhesus macaques. In all species, the vaccine elicited robust total IgG antibody responses against the full-length Spike, S1, S2, and receptor binding domain (RBD) proteins and strong T cell responses. Rhesus macaques (n=5/group) were intradermally administered two 1 mg doses of INO-4800 at a 4-week interval. Induction of memory immune responses was followed for several months following vaccination. The D614G SARS-CoV-2 Spike variant has emerged and is now dominant in >80% of isolates. INO-4800 vaccination induced neutralizing antibody responses against both the D614 and the now-dominant G614 SARS-CoV-2 Spike protein for >4 months. T cell responses were detected against the entire SARS-CoV-2 spike protein, with additional cross-reactive responses against SARS-CoV. Four months after vaccination, the INO-4800 vaccinated macaques and unvaccinated control animals were challenged with 1.1x104 PFU live SARS-CoV-2 virus (USA-WA1/2020) via intranasal and intratracheal inoculation. We observed memory recall of both antibody and T cell responses following viral challenge in vaccinated animals. These recall responses were associated with significantly lower viral loads in the bronchoalveolar lung lavage and faster nasal clearance compared to unvaccinated animals. This is the first study demonstrating vaccine-induced anamnestic protection against SARS-CoV-2 infection at a memory time point. Overall, the data support the induction of humoral and cellular immune responses by this SARS-CoV-2 DNA vaccine that are likely to have important impact on infection and support further study in people.
EX-VIVO MSC THERAPY FOR SEVERE COVID-19
Rita Barcia, PhD, Sentien Biotechnologies1, Brian O'Rourke, , Sunny Nguyen, , Arno Tellis, , Payal Garg, www.sentienbiotech.com2, Elisabeth LaPointe, , Chris Gemmiti, , Andrew Blair, , Brian Miller, , Biju Parekaddan
Sentien Biotechnologies1
www.sentienbiotech.com2
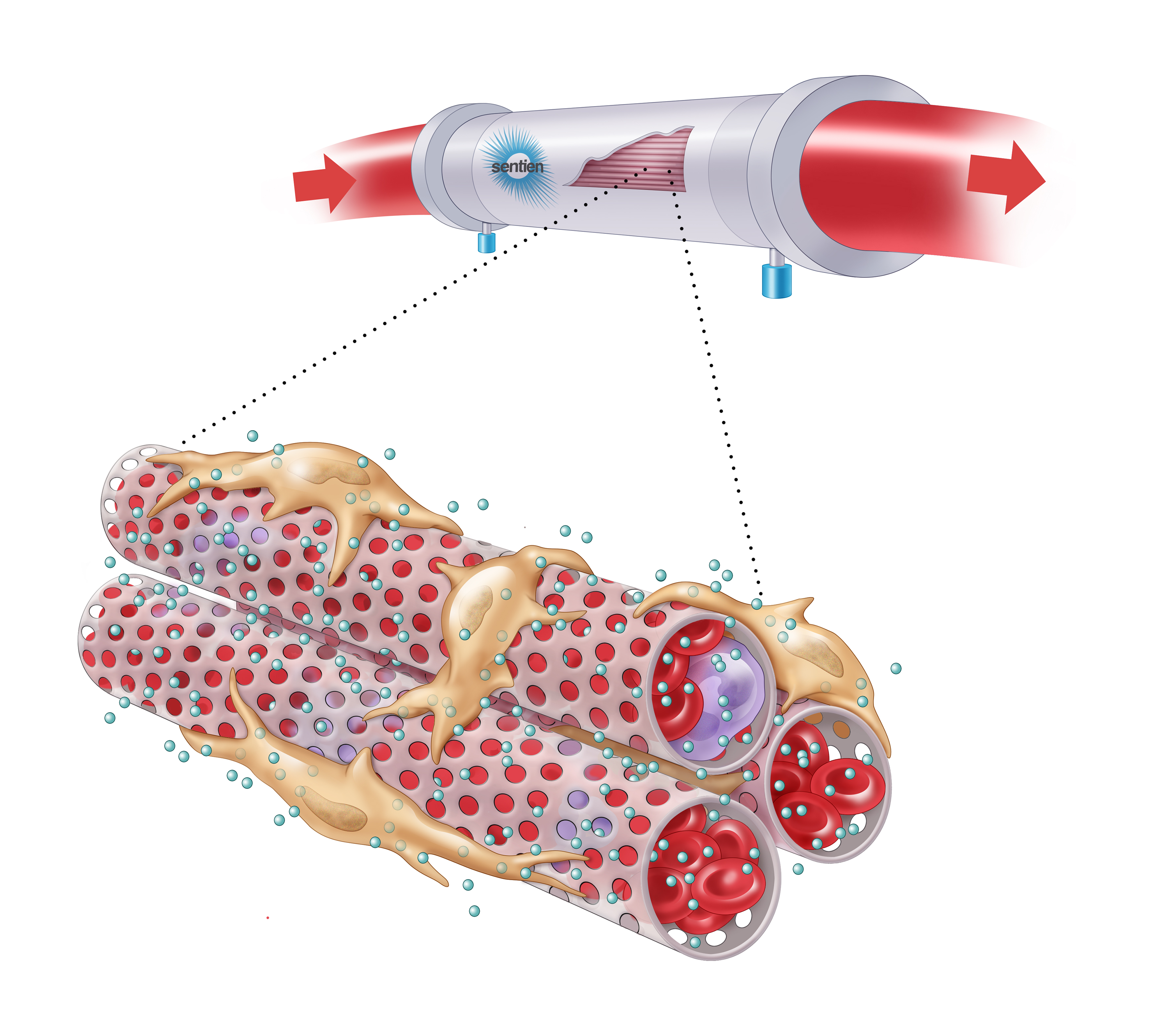
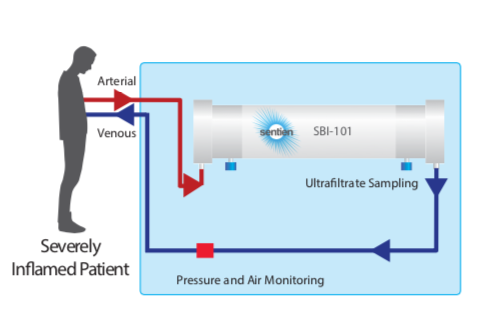
Mesenchymal stromal cells (MSCs) are a unique source of secreted factors that modulate an inflammatory response and enhance the repair of injured tissue. MSCs have been extensively studied in ARDS and other acute organ injuries. Sentien has created a novel delivery approach to enable sustained exposure to MSCs and their secreted factors, intended to overcome limits of cell transplantation/infusion while preserving their broad acting and dynamically responsive properties. Our lead product, SBI-101, contains allogeneic human MSCs inoculated into a hollow-fiber hemofilter, which enables communication with patient blood via the semi-permeable membrane, while maintaining MSC viability. Through this interplay, SBI-101 aims to restore balance to the immune system by reprogramming the molecular and cellular components of blood in patients with severe inflammation and organ injury. Sentien’s Phase I/II clinical study (NCT03015623) of SBI-101 in critically ill patients with Dialysis-Requiring Acute Kidney Injury (AKI-D) has produced data to support the therapeutic hypothesis of SBI-101 as a potent immunotherapy. Consistent with MSC biology, inflammatory markers, such TNFa and IFNg, were shown to be modulated, suggestive of a shift from a pro- to an anti- inflammatory state in treated patients. Data obtained in our AKI-D trial showed modulation of many biological molecules and immune populations that are significantly correlated with severe COVID-19 immunopathology. Here we make the case, using our existing AKI-D trial data, that SBI-101 may be of therapeutic benefit to severe cases of COVID-19. In addition we will present our study design for testing SBI-101 in COVID-19 patients, a multi-center, randomized, case-controlled, ascending-dose study in COVID-19 subjects with Acute Kidney Injury receiving Renal Replacement Therapy (NCT04445220) recently approved by the FDA.
Intravenously administered, AAV8-vectorized SARS-CoV2 monoclonal antibodies generate high level of serum neutralization titer in mice
Ye Liu, PhD, Regenxbio1, Xu Wang, REGENXBIO Inc.2, Devin McDougald, REGENXBIO Inc.2, Wei-Hua Lee, REGENXBIO23, Kirk Elliott, MS, REGENXBIO23, Chunping Qiao, PhD, REGENXBIO, Inc4, Timur Gaynutdinov, , Azadeh Bazarchi, , Ayda Mayer, , Joseph Bruder, , Olivier Danos, PhD, REGENXBIO23
Regenxbio1
REGENXBIO Inc.2
REGENXBIO23
REGENXBIO, Inc4
A novel coronavirus SARS-CoV2 has resulted in the worldwide COVID-19 pandemic since December 2019. The virus has caused tens of millions of confirmed infections and claimed more than half a million human lives. Multiple vaccines and therapeutics are in development to prevent or treat COVID-19. Among them, several SARS-CoV2 neutralizing antibodies have entered clinical trials for treatment or prevention. These antibodies have been isolated from recovered patients or transgenic mouse models and further engineered. They can bind to the receptor binding domain of the viral spike protein with high affinity and block viral entry into host cells through the ACE2 receptor. AAV vectorized antibody delivery has been shown to prevent viral infection from Ebola, Flu and HIV in rodent and NHP challenge models. Intramuscularly delivered AAV8 vectorized HIV antibody was shown to be well tolerated in Phase 1 human clinical trials. Phase 1/2 clinical trials for hemophilia, OTC deficiency and GSD1a involving intravenously delivered AAV8 and liver transgene expression have shown a positive safety and efficacy profile.
Liver targeted antibody expression has been shown to produce high level of serum antibody and induce immune tolerance in mouse and NHP models. In this study, we packaged the heavy and light chain coding sequence of three SARS-CoV2 monoclonal antibodies into the AAV8 capsid under the control of a liver specific enhancer and promoter. The antibodies are fully human IgG1 targeting the receptor binding domain of the spike protein. AAV8 vectors were intravenously administered to adult C57Bl6 mice at a dose of 1E13 vg/kg and serum human IgG levels were measured with ELISA after 1, 3, 5 and 7 weeks. Human IgG levels up to 1.4 mg/ml were reached in mouse serum at 3 weeks post vector administration and pVNT50 neutralization titers up to 1:500,000 were measured in a VSV-SARS-CoV2 pseudo-virus neutralization assay. Our work provides initial proof of concept for the feasibility of an AAV vectorized antibody approach to prevent COVID-19. It has the potential to serve as an alternate strategy to protect high-risk individuals or poor responders to vaccines. Additional dose finding study and SARS-CoV2 viral challenge study in animal models are warranted for further investigation.
Immune Cell Profiling After Treatment With High IL-6, A Marker Of Cytokine Release Syndrome In COVID19
Jamie Van Etten, PhD, Bio-Techne1, Greg Herr, , Christopher Hammerbeck, Bio-Techne Corporation2, Brian Astry, , Andrew Hudacek, , Jody Bonnevier, , Marnelle Andersen, , Kevin Flynn, Bio-Techne1
Bio-Techne1
Bio-Techne Corporation2
SARS-CoV2 infection and the resulting disease, COVID19, represent one of the largest public health challenges of the century. To date, over 17 million infections have been documented, resulting in over 600,000 deaths. COVID19 promotes activation of immune cells, including monocytes, dendritic cells, and macrophages, which leads to the upregulation of IL-8, interferons, TNF-alpha, and IL-6. In many patients, this hyperactivated immune response lead to systemic inflammation, tissue damage, and severe disease. Together, these symptoms are termed cytokine release syndrome. Importantly, elevated serum IL-6 has been associated with poor outcomes in patients with COVID19. Furthermore, numerous studies have observed a reduction in lymphocytes, notably NK and T cell populations, in patients with severe COVID19. We hypothesized that macrophages and monocytes partially contribute to IL-6 production in response to inflammatory cytokines, and that elevated IL-6 may dampen the cytotoxic response of T and NK cells.
Using high levels of IL-6 in cell culture media, we sought to mimic a hyperactivated immune response and measure its impact on primary immune cell growth and function. We expanded T cells or NK cells from peripheral blood mononuclear cell cultures for 9-14 days in serum-free culture medium with commercially available cytokines. Next, we measured the impact of IL-6 on T and NK cell expansion, as well as the functional impact of IL-6 in the culture media on T and NK cells. In addition, the neutralizing IL-6 antibody, sarilumab was assessed for its ability to rescue NK and T cell function. Using a multiplex immunoassasy platform, we determined the concentrations of a curated panel of secreted cytokines from these cell populations. We observed that IL-6 promoted proliferation of T cells and altered the secretion of various cytokines by T cells. Conversely, IL-6 had minimal impact on NK cell growth and function, as indicated by killing assays and analysis of cytokine secretion. Together, these data suggest that, while IL-6 signaling does not directly affect NK cell function, other cell types may be responsible for downregulation of NK cell function and growth. Overall, we observed IL-6 modulated T cell proliferation and function, which may be relevant for immune reactions in severe COVID19 disease.
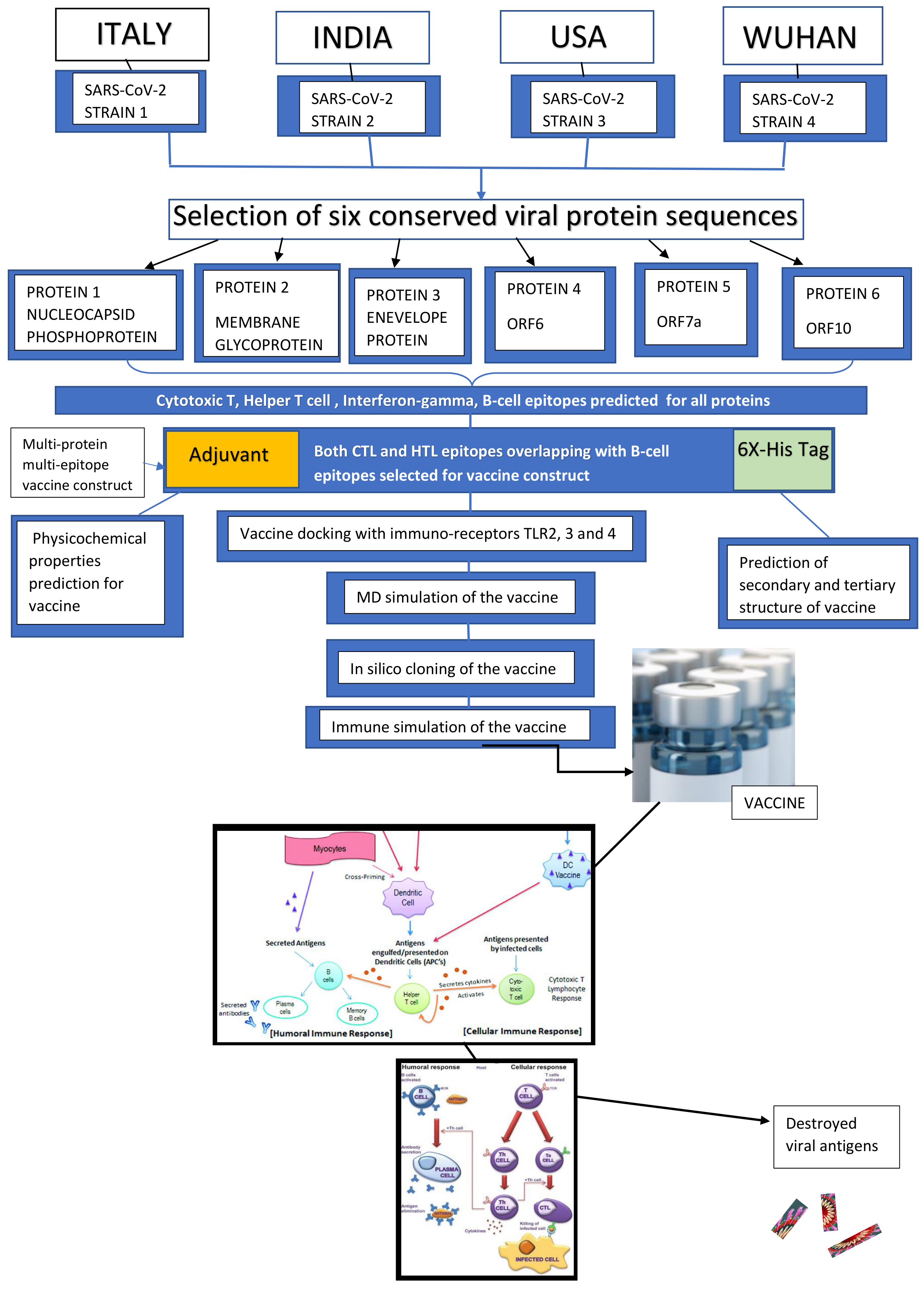
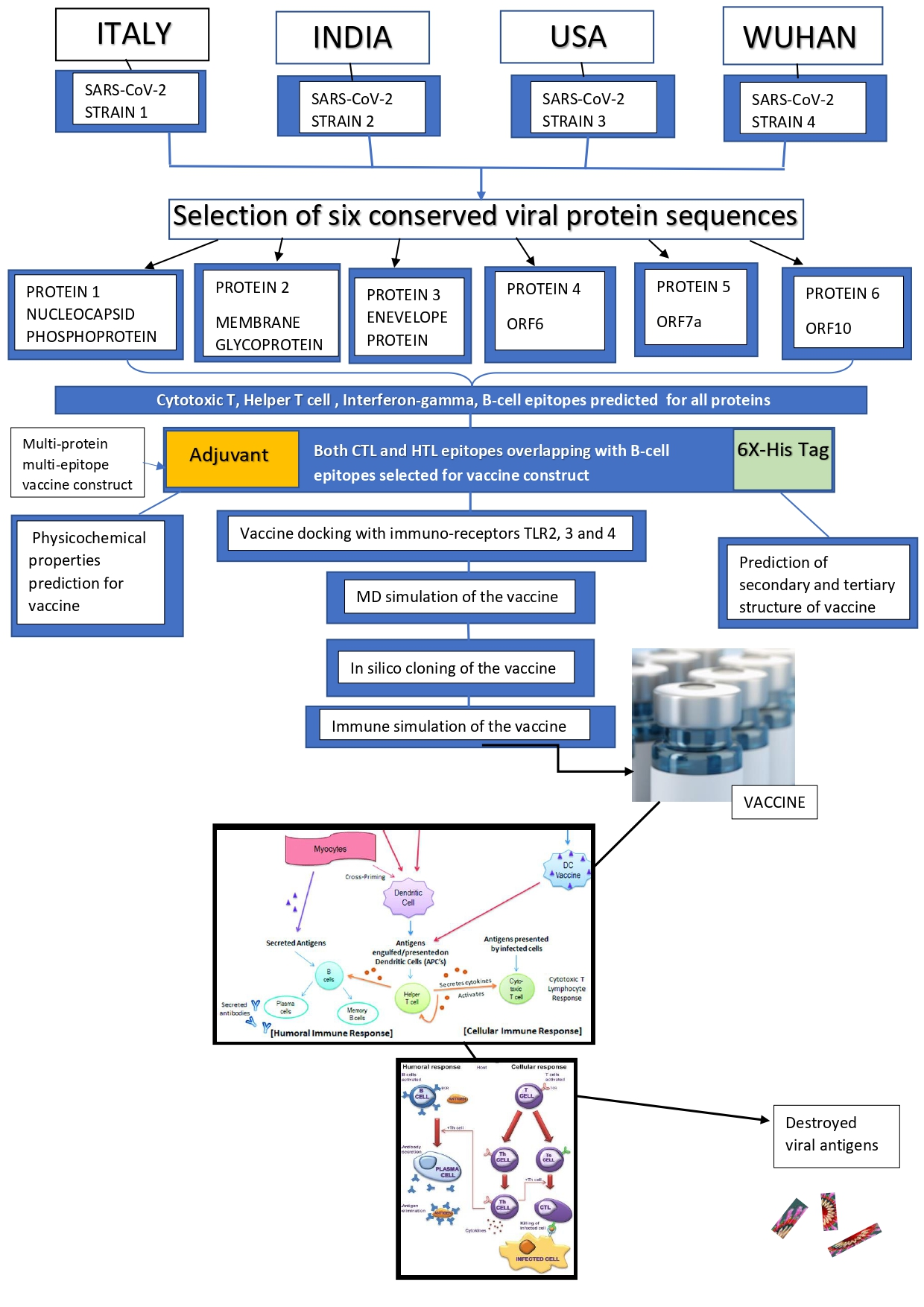
Immuno-informatics approach for multi-epitope vaccine designing against SARS-CoV-2
SOUVIK BANERJEE, MSc, ST. XAVIER'S COLLEGE (AUTONOMOUS), KOLKA1, Kaustav Majumder, Indian Institute of Technology Bombay2, Gerardo Gutierrez, Bachelor of Science, USF3, Debkishore Gupta, , Bharti Mittal
ST. XAVIER'S COLLEGE (AUTONOMOUS), KOLKA1
Indian Institute of Technology Bombay2
USF3
The novel Corona Virus Disease 2019 (COVID-19) pandemic has spread a blaze of increasing fatality rates across the world. The lack of effective vaccines has left the survival of mankind with doubts. The development of a multi-epitope vaccine in this current situation could be a possible treatment of COVID-19. We have designed a novel multi-epitope, multi-protein vaccine with different proteins of Severe Acute Respiratory Syndrome - Corona Virus -2 (SARS-CoV-2) using immuno-informatics approaches, which has been validated in silico to be stable and potential. It has been prepared with Cytotoxic T-cell (TC) and Helper T-cell (TH) binding epitopes overlapping with B-cell binding epitopes predicted for 6 proteins conserved among 4 different viral strains isolated across the world. Both the humoral and cell-mediated immune responses are ensured due to the presence of T cell and B-cell inducing epitopes along with interferon-gamma inducing epitopes present in the vaccine. The final vaccine construct comprises an adjuvant at the N terminal, Cytotoxic T Lymphocyte, and Helper T Lymphocyte epitopes. The construct showed potential antigenicity and was non-allergic. The molecular docking of the refined, validated tertiary structure model of the vaccine was performed with immune-stimulatory Toll-Like Receptors (TLR), TLR-2,3,4. The study of binding energetics of the docked complexes revealed binding interactions of receptors with the vaccine. Molecular dynamics simulation of the vaccine construct proves it to be stable within a biological system. The immune stimulation of the vaccine even confirmed the initiation of elevated host immune responses. The efficient translation of the vaccine in an expression vector was confirmed within silico cloning approach. Certainly, the development of such a vaccine candidate could possibly be an effective therapy for COVID-19.
Design of conditional small interfering RNA riboswitches to identify and selectively kill cells infected with SARS-CoV-2
Robin Hu, Undergraduate, University of Pennsylvania1, Saumya Das, MD, , John Rossi, PhD, Beckman Research Institute of City of Ho2, WILLIAM GODDARD, PhD, Caltech3
University of Pennsylvania1
Beckman Research Institute of City of Ho2
Caltech3
INTRODUCTION. 2,789,678 Americans have already been infected (as of July 4) with 129,305 deaths. These numbers are growing daily. Even with emergence of herd immunity or effective vaccines, SARS-CoV-2 will likely become endemic and continue to threaten vulnerable groups. SARS-CoV-2 is only the latest in a string of emergent diseases that have threatened human health. Since existing approaches are only partially effective, new therapeutic strategies are needed.
INNOVATION. To this end, we have developed novel conditional small interfering RNA riboswitches, Cond-siRNAs (Figure 1). Our riboswitch acts via toehold mediated displacement to hijack the RNA interference pathway and is active only in cells with the chosen disease biomarker. The identities of trigger and target genes for our riboswitch are encoded by two independent, easily programmable RNA sequences, giving the Cond-siRNAs the ability to target a specific cell population for the silencing of any arbitrary gene. Mechanistically, coupling of the SARS-CoV-2 mRNA transcript to our sensor releases the siRNA from the riboswitch to target BCL-2 family apoptosis inhibitors, inducing apoptosis of infected cells. In cells lacking the viral biomarker, there is no such activation. Thus, our RNAi drug is engineered to specifically target only virus-infected cells with minimal off-target effects. If coupled with other early detection strategies, our therapy may target even mildly symptomatic or asymptomatic patients.
RESULTS. The sensor “trigger” gene candidates of our COVID-19 riboswitch were taken from all possible 31 nucleotide segments in the 195 highly conserved regions of the SARS CoV-2 sequence. After ranking sensor sequences for uniqueness in the human transcriptome and modeling thermodynamic stability scores, we optimized placement of LNA chemical modifications. Through this iterative process, we have identified and designed eight unique Cond-siRNAs, all of which demonstrate potential from a structural standpoint (Figure 2). We intend to test all eight using a reporter that carries a SARS-CoV2 sequence (but not the full virus itself) to demonstrate its ability in silencing the chosen target in pilot Caco-2 cell culture studies. Success in these proof-of-concept experiments would escalate towards testing the full virus in a Caco-2 viral replication assay.
CONCLUSION. We believe this strategy addresses an unmet need to safely treat asymptomatic or minimally symptomatic patients, a population that has been the lynchpin of community spread. Our programmable Cond-siRNA riboswitch allows for post-delivery targeting of RNAi activity to specific disease-presenting cells, compatible and practical for development into clinically viable RNAi drugs. The immediate goal is to stop the current pandemic, but a continuous strategy for future applications is needed. Our strategy requires only sequence information and could provide rapid responses to tackle the next threat before it becomes pandemic.

Safety and immunogenicity of VSV-SARS2 as a potential COVID19 vaccine: Preliminary findings in cynomolgus macaques
Melanie Graham, PhD, , Patrycja Lech, PhD, , Mulu Tesfay, PhD, Imanis life sciences1, Clement Gnanadurai, PhD, , Rianna Vandergaast, PhD, Imanis Life Sciences1, Timothy Carey, PhD, Imanis Life Sciences1, Samantha Reiter, , Christopher Petro, PhD, , Chase Lathrum, , Toshie Sakuma, PhD, , Timothy Carey, PhD, Imanis Life Sciences1, Luke Russell, PhD, Vyriad2, Bethany Brunton, PhD, Vyriad2, Jordan Recker, Vyriad2, Lukkana Suksanpaisan, PhD, , Justin Koepsel, PhD, Vyriad2, Alina Baum, PhD, , Christos Kyratsous, PhD, , Stephen Russell, MD, PhD
Imanis life sciences1
Vyriad2
VSV-SARS2 is a recombinant, replicating Indiana strain vesicular stomatitis virus encoding the SARS-CoV-2 spike glycoprotein (GP) in place of the native VSV attachment glycoprotein (VSV.G) and might have utility as a vaccine for COVID19. Six healthy male cynomolgus macaques age 3.5 to 4.4 years were equally randomized to three groups: 108 TCID50 of a VSV-SARS2 preparation displaying both VSV.G and spike GP was administered intramuscularly (IM) or orally (PO) or 107 TCID50 of a VSV-SARS2 preparation with only spike GP given PO. Animals were monitored closely for toxicity, viremia, virus shedding in urine and saliva, and antibody response to the SARS-CoV2 spike glycoprotein on days 1, 4, 8, 11, 14, 21, 28 and 42. Body temperature was mildly elevated during short-term follow-up (101.3±0.8°F) compared with baseline (98.6±1.8°F), and in 5 of the 6 animals, Grade 1 mucositis was observed but did not interfere with normal daily activities and resolved without treatment. Episodic vomiting unrelated to the vaccine was observed, and was related to the sedation that was given to enable test article administration and sampling. Viremia was detected day 1 in both of the animals vaccinated by the IM route, but not at later time points and was never detected in orally vaccinated animals. Virus shedding in urine, saliva, feces, buccal, or nasal swabs was negative by PCR at all timepoints tested in all animals, and no infectious virus was detected in any rectal, buccal or nasal swabs from any animal.
SARS-CoV-2 neutralizing antibodies were evaluated using a pseudotype neutralization assay and confirmed using a SARS-CoV-2 clinical isolate PRNT assay. Neutralizing antibodies were first detected on day 8 post vaccination and at all subsequent timepoints in 2/2 animals following IM administration and on day 11 post vaccination and at all subsequent timepoints in 2/4 animals following PO administration, independent of VSV.G. All four animals with detectable neutralizing antibodies showed parallel increases in their IgG and IgM antibody titers against immobilized fragments of the SARS-CoV-2 spike glycoprotein (S1/S2, S1 only, and RBD) and against the trimer form. Also, both of the IM vaccinated animals, but none of the orally vaccinated animals, developed anti-VSV G antibodies capable of neutralizing wild type VSV.
On day 42 post vaccination, the two orally vaccinated animals that had failed to seroconvert were vaccinated by IM injection of 107 or 105 TCID50 of the VSV-SARS2 virus. Both of these animals developed SARS-CoV-2 neutralizing antibodies within 14 days of the redosing.
In summary, VSV-SARS2 demonstrated a favorable safety profile and appears to be a promising candidate for clinical evaluation as a SARS2 Coronavirus vaccine.
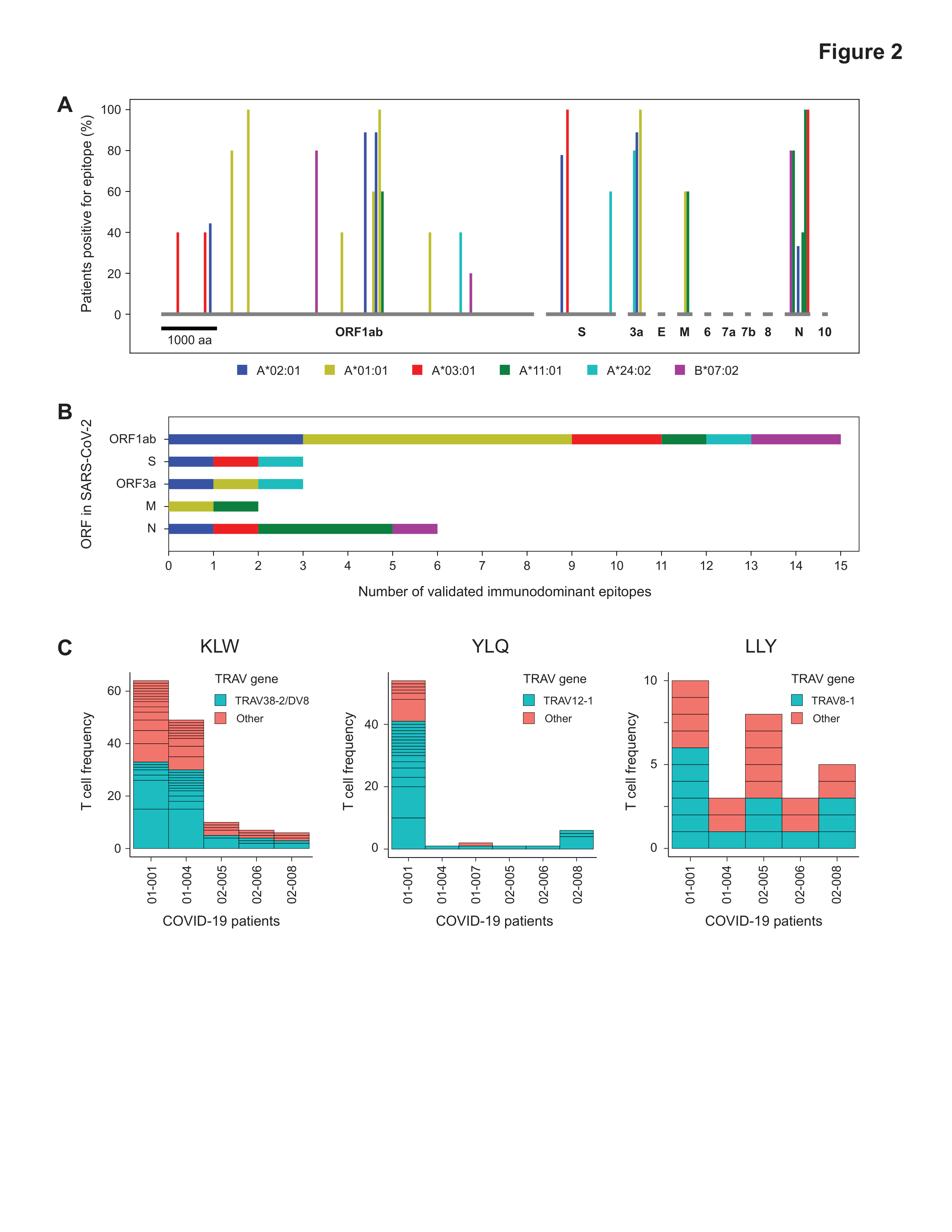
UNBIASED GENOME-WIDE DISCOVERY USING TSCAN REVEALS SHARED IMMUNODOMINANT CD8+ T CELL EPITOPES IN SARS-COV-2
Gavin MacBeath, PhD, TScan Therapeutics1, Andrew Ferretti, , Tomasz Kula, , Yifan Wang, , Dalena Nguyen, , Adam Weinheimer, , Garrett Dunlap, , Qikai Xu, , Nancy Nablisi, , Candace Chouinard, , Alexander Cristofaro, , Holly Whitton, , Amy Virbasius, , Ken Olivier, Tscan2, Lyndsey Buckner, , Angela Alistar, , Eric Whitman, , Sarah Bertino, , Shrikanta Chattopadhyay
TScan Therapeutics1
Tscan2
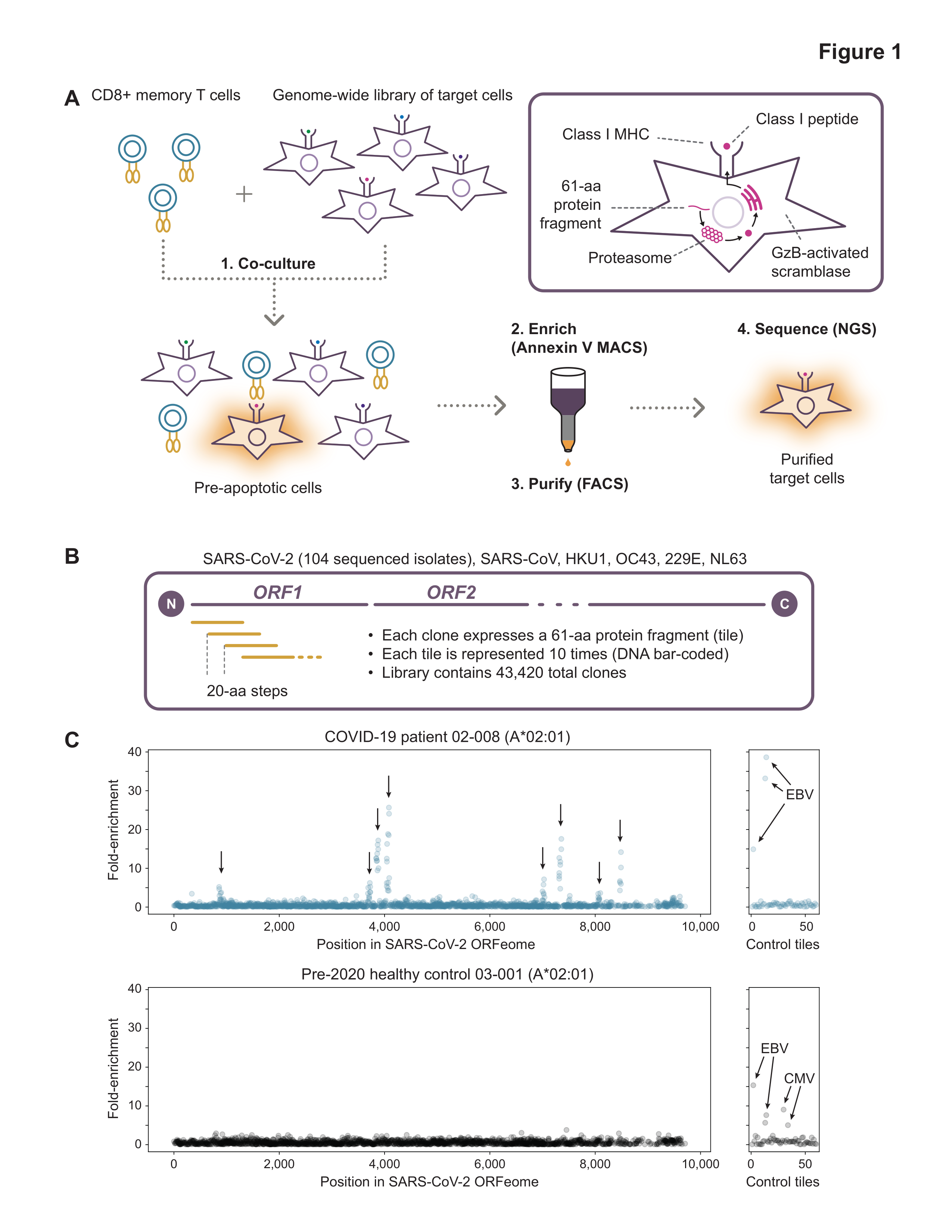
Development of effective strategies to detect, treat, or prevent COVID-19 requires a robust understanding of natural immunity to SARS-CoV-2, including the cellular response mediated by T cells. We used an unbiased, genome-wide screening technology, termed T-Scan, to comprehensively identify the specific epitopes in SARS-CoV-2 that are recognized by the memory CD8+ T cells of 25 COVID-19 convalescent patients. For each of six HLA types examined, patient T cells recognized 3–8 immunodominant epitopes that are broadly shared among patients, and single-cell sequencing revealed common structural features of TCRs recognizing these epitopes. We detected minimal cross-reactivity to the endemic coronaviruses that cause the common cold, arguing that pre-existing immunity to other coronaviruses does not significantly shape CD8+ T cell responses to SARS-CoV-2. Notably, only 3 of the 29 immunodominant epitopes we identified reside in the Spike protein, highlighting the need for second-generation vaccines that recapitulate natural CD8+ T cell immunity to SARS-CoV-2.
SARS-CoV-2 infects human neural progenitor cells and brain organoids
Hin Chu, , Baozhong Zhang, PhD, , Kwok-yung Yuen
SARS-CoV-2 infection primarily causes respiratory illness with clinical manifestations largely resembling those of SARS. However, neurological symptoms have also been frequently reported in COVID-19 patients, and SARS-CoV-2 RNA has been detected in brain biopsies of fatal COVID-19 cases. Despite these clinical observations, so far there has been no direct experimental evidence of SARS-CoV-2 infection in the human central nervous system (CNS). In this study, we investigated the infection and replication of SARS-CoV-2 in iPSC-derived human neural progenitor cells (hNPCs), neurospheres, and human brain organoids. Our study revealed a number of important findings:
(1) SARS-CoV-2, but not SARS-CoV, could infect and replicate in hNPCs.
(2) SARS-CoV-2 productively infected neurospheres and human brain organoids with release of infectious virus particles.
(3) SARS-CoV-2 was identified in TUJ1- and NESTIN-positive cells, suggesting the virus could target cortical neurons and neuronal progenitor cells in human brain organoids.
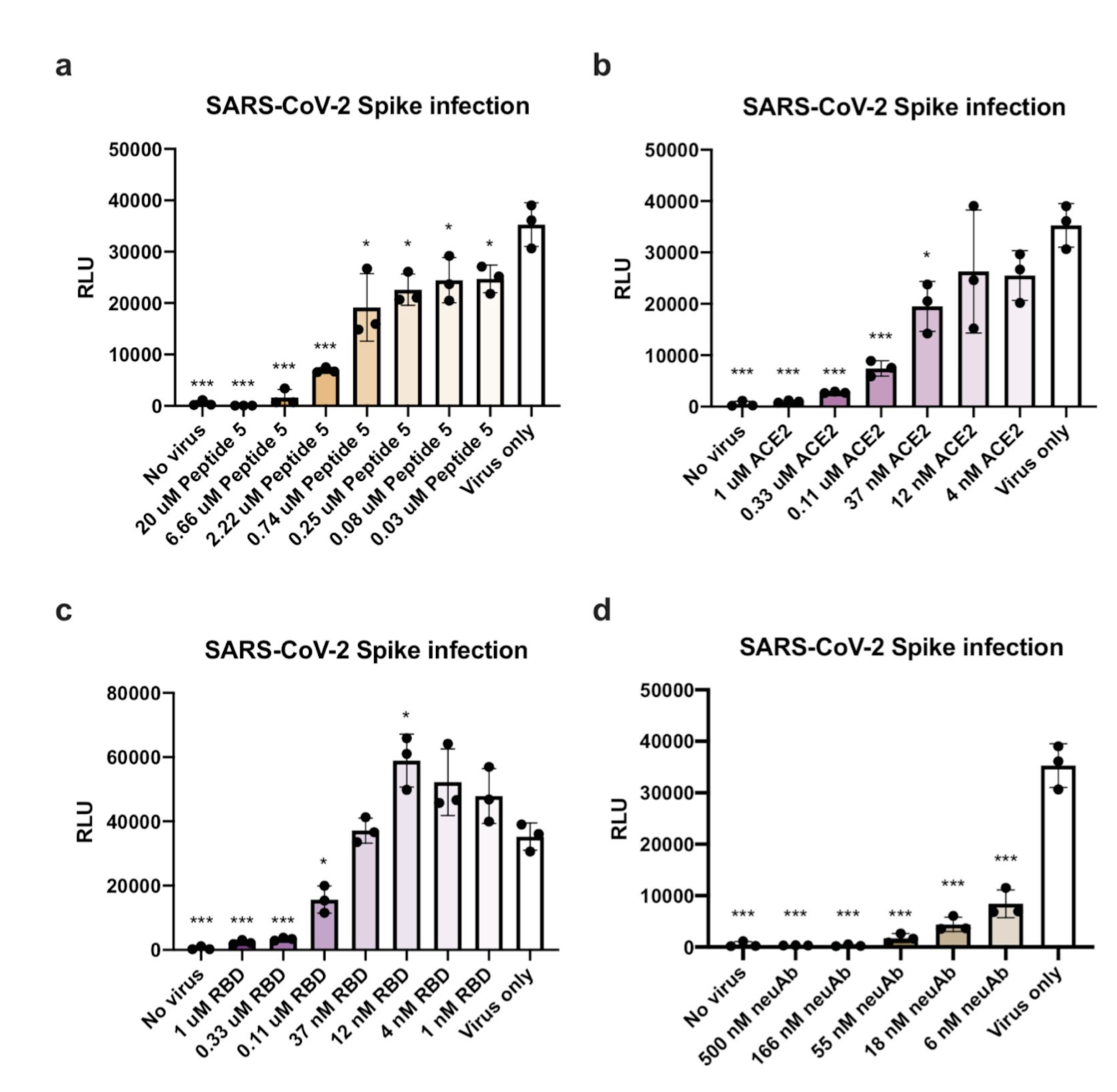
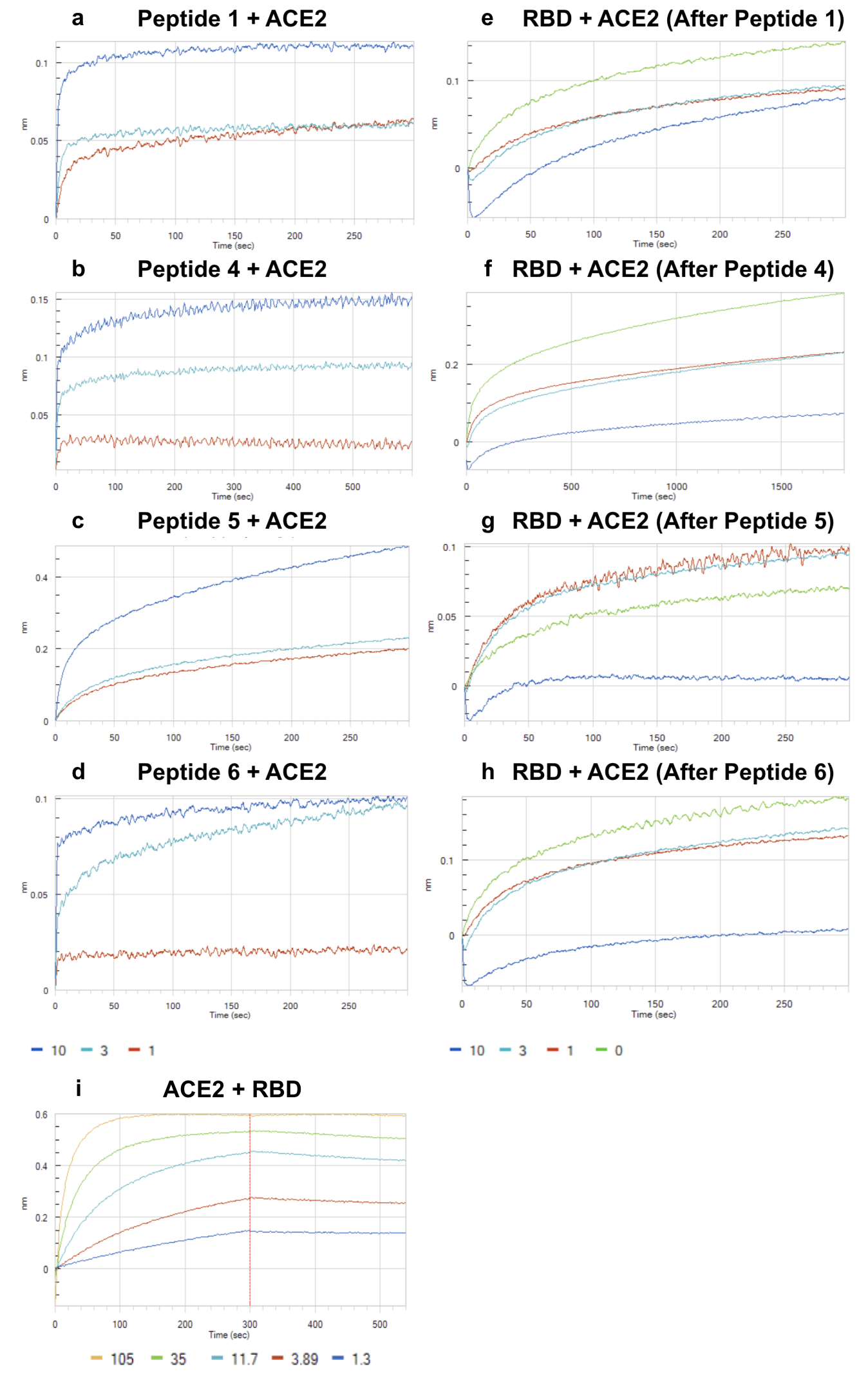
Peptide Antidotes to SARS-CoV-2 (COVID-19)
Andre Watson, BS, Ligandal Inc.1, Leonardo Ferreira, PhD, University of California San Francisco2, Peter Hwang, PhD, , Jinbo Xu, PhD, , Robert Stroud, PhD
Ligandal Inc.1
University of California San Francisco2
The design of an immunogenic scaffold that serves a role in treating a pathogen, and can be rapidly and predictively modeled, has remained an elusive feat. Here, we demonstrate that SARS-BLOCK™ synthetic peptide scaffolds act as antidotes to SARS-CoV-2 spike protein-mediated infection of human ACE2-expressing cells. Critically, SARS-BLOCK™ peptides are able to potently and competitively inhibit SARS-CoV-2 S1 spike protein receptor binding domain (RBD) binding to ACE2, the main cellular entry pathway for SARS-CoV-2, while also binding to neutralizing antibodies against SARS-CoV-2. In order to create this potential therapeutic antidote-vaccine, we designed, simulated, synthesized, modeled epitopes, predicted peptide folding, and characterized behavior of a novel set of synthetic peptides. The biomimetic technology is modeled off the receptor binding motif of the SARS-CoV-2 coronavirus, and modified to provide enhanced stability and folding versus the truncated wildtype sequence. These novel peptides attain single-micromolar binding affinities for ACE2 and a neutralizing antibody against the SARS-CoV-2 receptor binding domain (RBD), and demonstrate significant reduction of infection in nanomolar doses. We also demonstrate that soluble ACE2 abrogates binding of RBD to neutralizing antibodies, which we posit is an essential immune-evasive mechanism of the virus. SARS-BLOCK™ is designed to “uncloak” the viral ACE2 coating mechanism, while also binding to neutralizing antibodies with the intention of stimulating a specific neutralizing antibody response. Our peptide scaffolds demonstrate promise for future studies evaluating specificity and sensitivity of immune responses to our antidote-vaccine. In Figure 1a-1d, we show SARS-CoV-2 pseudotyped lentiviral infections of ACE2-expressing cells inhibited by Peptide 5 (a), ACE2 (b), RBD (c), and neutralizing antibody (d), with Peptide 5 exhibiting ~30nM potency and blocking 95% of infection at 6.6uM dose. In Figure 2a-2d, peptides 1, 4, 5 and 6 were associated with ACE2 at 1, 3 and 10μM concentrations until saturation was observed. Next, in Figure 4e-4H, we demonstrated that peptides 1, 4, 5 and 6 are able to inhibit SARS-CoV-2 binding to ACE2. In summary, SARS-BLOCK™ peptides are a promising COVID-19 antidote designed to combine the benefits of a therapeutic and vaccine, effectively creating a new generation of prophylactic and reactive antiviral therapeutics whereby immune responses can be enhanced rather than blunted.
Repurposing Generic Drugs Against COVID-19: Potential Binders of Viral Targets Nibedita Rath1 1Open Source Pharma Foundation, National Institute of Advanced Studies, IISc Campus, Bangalore, India
Nibedita Rath, PhD, Open Source Pharma Foundation1, Jaykumar Menon
Open Source Pharma Foundation1
The outbreak of COVID-19, a new strain of coronavirus was emerged from Wuhan in late 2019, China and has spread globally ever since it's outbreak. Currently, there is no definite treatment for COVID-19, although few therapeutics, such as small molecules, vaccines, antibodies are under investigations. The need of the hour is to come up with a treatment at the earliest possible. So, our strategy is to look into generic drugs that could be repurposed against COVID-19. Identifying new uses for off patent drugs would make the drug discovery process much faster, cheaper and less risky than developing new drugs and therefore offers what may be the single most promising avenue for delivering new medical treatments to the current pandemic. The current work has tried to identify potential binder of SARS-CoV-2 Mpro main protease enzyme (Mpro,3CLpro), substrate binding pocket. The most fetching drug target of Covid-19 is the main protease (Mpro,3CLpro, Nsp5) due to its imperative role in processing the polyproteins that are translated from the viral RNA. The structure based virtual screening of 1900 generic drugs followed by identification of hits based on their calculated binding energies, poses and interactions with key amino acids. The top hits include antiviral drugs some of which are under investigation in clinic for Covid-19. A few of the most promising hits in our screen are the drugs Leuprolide, Rutin, Caspofungin, Spiramycin, Betamethasone, Saquinavir and Ginseng that are not reported as potential options to the best of our knowledge. In addition, the top hits bound to the terminal site of Mpro substrate-binding pocket include the antibiotics drug and vasodilator drugs among others. These results can be used as a starting point for further in vitro and in vivo testing, either individually or in combinations and followed by testing in a clinical trial setup.
Development and Validation of a High-Throughput Clinical Assay for Detecting SARS-CoV-2-Neutralizing Antibodies
Rianna Vandergaast, PhD, Imanis Life Sciences1, Timothy Carey, PhD, Imanis Life Sciences1, Samantha Reiter, , Patrycja Lech, PhD, , Clement Gnanadurai, PhD, , Chase Lathrum, , Ryan Johnson, , Mulu Tesfay, PhD, Imanis life sciences1, Jason Buehler, PhD, Imanis Life Sciences1, Lukkana Suksanpaisan, PhD, , Shruthi Naik, PhD, , Bethany Brunton, PhD, Vyriad2, Jordan Recker, Vyriad2, Michelle Haselton, , Christopher Ziegler, PhD, , Anne Roesler, , John Mills, PhD, , Elitza Theel, PhD, , Scott Weaver, PhD, , Grace Rafael, , Matthew Roforth, , Calvin Jerde, , Sheryl Tran, Vyriad2, Rosa Maria Diaz, PhD, , Alice Bexon, MD, , Alina Baum, PhD, , Christos Kyratsous, PhD, , Kah-Whye Peng, PhD, Vyriad2, Stephen Russell, MD, PhD
Imanis Life Sciences1
Vyriad2
We have recently developed and validated IMMUNO-COVTM, a high-throughput clinical test to quantitatively measure SARS-CoV-2-neutralizing antibodies, the specific subset of anti-SARS-CoV-2 antibodies that block viral infection. The test measures the capacity of serum or plasma to neutralize virus infection using a replicating recombinant Vesicular Stomatitis Virus (VSV-SARS-CoV-2-S-d19CT) as a safe surrogate to SARS-CoV-2. VSV-SARS-CoV-2-S-d19CT induces fusion in Vero cell monolayers, which is detected in the assay as luciferase signal using a dual split protein (DSP) reporter system. Purified SARS-CoV-2 neutralizing antibodies and plasma or serum from SARS-CoV-2 convalescing individuals blocked VSV-SARS-CoV-2-S-d19CT infection, resulting in a measurable reduction in luciferase signal. During validation and verification studies, the assay exhibited 100% specificity. In blinded analyses, assay results demonstrated near-perfect correlation (196/197) with available clinical data and qRT-PCR or other serological testing results. We generated a calibration curve consisting of stepped concentrations of α-SARS-CoV-2-spike monoclonal antibody added to pooled SARS-CoV-2 seronegative serum or plasma matrix. By using the calibration curve, the magnitude of the SARS-CoV-2-neutralizing response in test samples was quantitated from a single test sample dilution. For samples with high levels of SARS-CoV-2-neutralizing antibodies, a second dilution facilitated more precise quantitation. The virus neutralization units (VNUs) calculated using this calibrator method correlated closely (p < 0.0001) with plaque reduction neutralization titer-EC50 (PRNTEC50) values determined by plaque reduction neutralization test against a clinical isolate of SARS-CoV-2. Thus, this surrogate neutralization assay accurately measures SARS-CoV-2-neutralizing antibodies in a BSL2, high-throughput format, making it a valuable addition to the other currently available serological tests. In particular, the assay can provide vital information for evaluating donor eligibility for convalescent plasma therapy programs and participant eligibility for various clinical trials, as well as assessing efficacy in terms of immune responses to candidate SARS-CoV-2 vaccines in development. The assay is also commercially available in the USA.
ISOTHERMAL AMPLIFICATION AND DETECTION OF SARS-COV-2 USING CRISPR TECHNOLOGY
Gabriel Lamothe, Msc student, Laval University1, Jacques Tremblay, PhD, Université Laval and CRCHUQ2
Laval University1
Université Laval and CRCHUQ2
Background: The SARS-CoV-2 viral infection has plagued humanity since the end of December 2019. It has been considered the newest and biggest worldwide threat. Of the many methods that have been developed and adapted to contain the pandemic, detection tests are on the frontlines. However, the current reliance on old, PCR-based technologies means that these tests are limited to areas with highly trained personnel and laboratory equipment such as thermocyclers. While this is less of an issue in developed countries with a strong biotechnology industry and academic environment, this has proven challenging for less developed countries. We have therefore worked on adapting the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK)-based viral RNA detection system to meet these new needs.
Methods: SHERLOCK is based on viral RNA detection system. The major component of this system is based on the collateral cleavage capabilities of Cas13a. Upon recognizing its target RNA, Cas13 is activated and begins cleaving all surrounding RNAs. To ensure that the Cas13 is capable of producing a signal of great enough intensity, a three steps process is used to amplify a section of the original SARS-CoV-2 genome. First, the virus RNA is reverse transcribed to create a cDNA that can be amplified by Loop-mediated isothermal amplification (LAMP). The original SHERLOCK technology used Recombinase Polymerase Amplification, however, due to the limited supply chain for the required enzymes, we have shifted our focus to LAMP. To ensure transcription can proceed smoothly after the amplification, a T7 promoter insert is included in the FIP primer. We are currently performing this on the SARS-CoV-2 N-gene control plasmid supplied by IDT. This is leveraged to produce RNA targets for the Cas13a-based detection. The Cas13a is provided with a crRNA that can be programmed to hybridize to specific sequences of the SARS-CoV-2 genome and result in the activation of the enzyme.
Results: As proof of concept, we have achieved a strong LAMP amplification with a T7 promoter included in the FIP primer. This demonstrates that this isothermal amplification process can be leveraged in our detection test. Additionally, we have used our Cas13a protein in conjunction with RNAse ALERT from IDT to create a visual readout. When incubated with a crRNA and its associated target RNA, Cas13a is activated and begins its collateral cleavage process. We have used this to cleave the reporter RNA in RNAse ALERT and separate the fluorescent molecule from its quencher. Under a black light, it is possible to see which samples have been properly cleaved.
Conclusion: We have shown that this new adaptation to SHERLOCK has potential in detecting SARS-CoV-2 with a decreased reliance on laboratory equipment.
Therapeutic efficacy of human umbilical cord mesenchymal stem cells for severe COVID-19 patients
Zhan Li, Dr, Army Medical University1, Wei Xing, , Dongpo Jiang, , Xiang xu
Army Medical University1
COVID-19 was the disease caused by the novel SARS-CoV-2 coronavirus, which could make many patients become critically ill with severe pneumonia. COVID-19 is potentially a life-threatening condition and a major cause of death in the severe and critically ill patients, which is characterized by multiple organ dysfunctions as a result of unbalanced host inflammatory response to pathogens. For the COVID-19 patients, there appears to be a frequently observed pathology-a cytokine storm, damaged organs (lung, liver, kidney, heart), reduced lymphocyte and increased D-dimer. However, effective treatment or intervention to prevent COVID-19 associated morbidity is lacking. Human umbilical cord mesenchymal stem cells (hUC-MSCs) transplantation was considered as a promising approach for COVID-19 treatment because of its tissue repair and potent immunomodulatory properties. Taken together with our previous study of hUC-MSCs therapy for severe sepsis patients, we make a further research and exploration in the efficacy of hUC-MSCs treatment in severe COVID-19 patients at Huoshenshan hospital. Our team enrolled 20 patients (severe illness, Age > 18 years) who averagely divided into MSCs treatment plus standard of care and only standard of care cohorts. hUC-MSCs were administered to ten COVID-19 patients by intravenous injections (4×107 cells per people in 100ml normal saline) each on days 1, 3 and 6. During the 30-day post-infusion period, we performed a series of tests to evaluate the safety or efficacy of MSC administration associated with the standard of care. Our results indicated that administration of hUC-MSCs significantly improved pulmonary fibrosis, increased lymphocyte and reduced D-dimer in severe COVID-19 patients. Our data also suggested that the patient's vital signs were stable, and no safety concerns were identified.
IN SILICO DESIGN OF NOVEL COVID-19 DRUG THROUGH STRUCTURE BASED VIRTUAL SCREENING FOLLOWED BY VALIDATION USING THE FLOWER TECHNIQUE WITH ATTOMOLAR SENSITIVITY
Soo-Kyung Kim, PhD, , Judith Su, PhD, , WILLIAM GODDARD, PhD, Caltech1
Caltech1
The emergence and rapid spread of a novel severe acute respiratory syndrome (SARS)-like coronavirus SARS-CoV-2 is destroying global health and economy.(Li, Guan et al. 2020) This virus forces much of the world to adopt a lockdown mode, causing staggering economic fallout and human suffering (https://www.cdc.gov/coronavirus/novel-coronavirus-2019.html). Already, SARS-CoV-2 has infected over 17 million people and caused more than ~666,000 deaths (https://www.worldometers.info/coronavirus/).
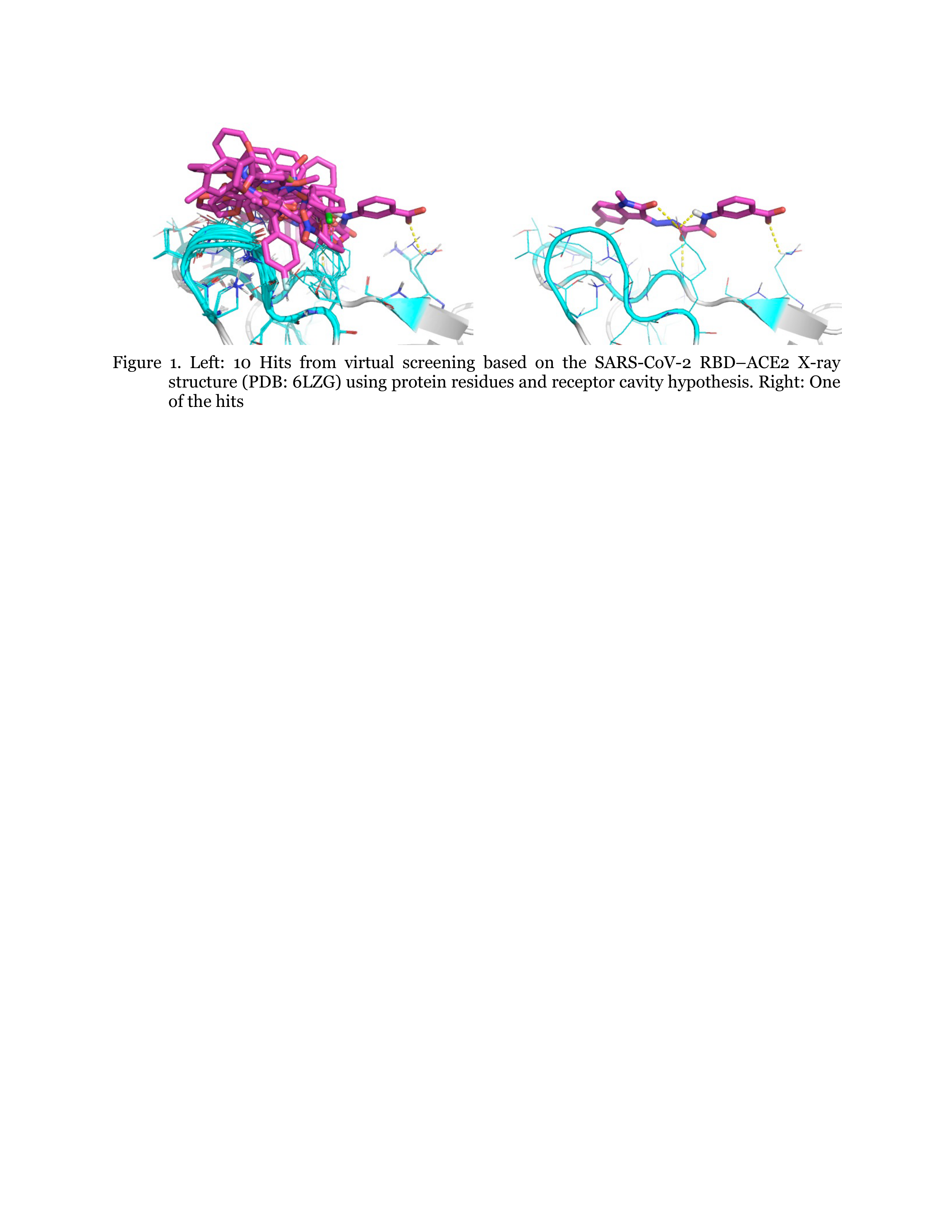
We performed in silico lead discovery computational methods to identify novel small molecule inhibitors that bind to the Receptor-Binding Domain (RBD) of SARS-CoV-2 spike, thereby preventing interaction with human angiotensin-converting enzyme 2 (hACE2), the first step of virus entry. For virtual screening (VS), we used the ZINC FDA approved Drug Bank including ~1,657 drugs. (http://zinc15.docking.org/catalogs/dbfda/). We generated a pharmacophore with protein residue hypothesis using the 2.5-Å X-ray structure of SARS-CoV-2 complexed with hACE2 (PDB ID: 6LZG). We found 46 hit molecules from VS, some of which are shown in Figure 1. We identified 2 known drugs that we predict will bind strongly to the RBD. Using the DarwinDock complete sampling method, we predicted improved ligand binding poses, showing strong salt-bridges at R403, R408, and K417.
To validate experimentally the binding of these 2 drugs to the RBD, we will use the frequency-locked optical whispering gallery mode evanescent resonator (FLOWER) system. See Figure 2. This system has previously been demonstrated to have attomolar sensitivity. A main advantage of FLOWER is that because of its high sensitivity it is not necessary to label the target compounds. Also only small sample volumes are required. As such, these sensors provide an ideal drug discovery platform. To perform these experiments, we covalently bind anti-RBD IgG antibodies to the surface of the silica microtoroid. We generate a dose response curve and obtain the binding affinities.
Since the molecules identified in this study have already advanced into the clinic, the known pharmacological and human safety profiles of these compounds will enable accelerated preclinical and clinical evaluation of these drugs for the treatment of COVID-19. Success could have enormous impact on the treatment of patients suffering from SARS-CoV-2 and on our approach to treating other SARS-associated coronavirus.

Preclinical Efficacy Evaluation of a DNA Vaccine Ttargeting SARS-COV-2 in a hACE2 Transduced Mouse Challenge Model
Ebony Gary, PhD, The Wistar Institute1, Bryce Warner, PhD, , Elizabeth Parzych, PhD, , Bryan Griffin, PhD, , Xizhou Zhu, , Nikesh Tailor, , Nicholas Tursi, , Mable Chan, , Mansi Purwar, , Robert Vendramelli, , Emma Reuschel, PhD, , Yanlong Pei, , Kevin Liaw, Phd, , Sylvia Thomas, University of Guelph2, Edgar Tello, , Ali Ali, , Matthew Guilleman, Bsc, MBIOT, University of Guelph2, Amira Rghei, University of Guelph2, Sarah Wootton, PhD, University of Guelph2, Ami Patel, PhD, The Wistar Institute1, Trevor Smith, PhD, , Kar Muthumani, PhD, , David Weiner, PhD, The Wistar Institute1, Darwyn Kobasa, PhD
The Wistar Institute1
University of Guelph2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in the human population in late 2019 and is the causative agent of coronavirus disease of 2019 (COVID-19). To date, more than 17 million people have been infected with SARS-CoV-2 and COVID-19 has caused over 500,000 deaths worldwide. We developed a DNA vaccine encoding the SARS-CoV-2 spike glycoprotein (INO-4800) which has since entered clinical trials (NCT04336410, NCT04447781). As wild-type mice do not express angiotensin-converting enzyme 2 (ACE2) which serves as the receptor for the SARS-CoV-2 spike glycoprotein, easily accessible mouse models of SARS-CoV2 infection are limited. Here, we characterized immune responses to a similar DNA vaccine antigen encoding full-length SARS-CoV-2 spike glycoprotein (pS), and employed a mouse model based on transduction of the respiratory tract of wild type BALB/c mice with a lung tropic adeno-associated virus vector (AAV6.2FF) carrying the gene for human ACE-2 (AAV6.2FF-hACE2) to evaluate vaccine efficacy in vivo. Immunization with pS induced robust spike-specific T cell responses as measured by IFNy ELISpot and intracellular cytokine staining. pS immunization resulted in the rapid development of robust anti-spike IgG titers in the serum of immunized mice. IgG2a was the predominant isotype induced by pS immunization and resulted in an increased IgG2a:IgG1 ratio. pS-immunization elicited antibody responses that persisted more than 100 days post-final immunization. Serum from pS-immunized mice neutralized SARS-CoV-2 spike-pseudotyped viruses in vitro. Finally, pS immunized mice transiently expressing hACE2 had decreased infectious SARS-CoV-2 virus titers and reduced viral RNA in their lungs as compared to unimmunized after challenge with SARS-CoV-2. These data demonstrate that spike-encoding DNA vaccines are immunogenic and protective in vivo, support the continued translation of these constructs to the clinic, and demonstrate that the AAV6.2FF-hACE2 mouse transduction model represents an easily accessible small-animal model for wild-type SARS-CoV-2 infection which can be used to evaluate anti-SARS-CoV-2 vaccine efficacy.
Establishing A Platform For High Titre SARS-CoV-2 S Pseudotyped Lentiviral Vectors – A Tool For Anyone To Play With
Kamran Miah, PhD, University of Oxford1, Yue Du, PhD, University of Oxford1, Dwiantari Satyapertiw, , Rebecca Dean, , Catriona Conway, University of Oxford1, Stephen Hyde, PhD, University of Oxford1, Deborah Gill, PhD, University of Oxford1
University of Oxford1
Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV 2), responsible for the current COVID-19 pandemic, has resulted in over 18 million cases globally with significant fatality rates. The need for SARS-CoV-2-related therapeutics (chemotherapies, immunoprophylaxes, and vaccines) is highly stressed by the World Health Organisation. However, the capabilities for many research groups to contribute to COVID-19 research is limited by the tight biological safety level (BSL) requirements for working with authentic SARS-CoV-2 virus (BSL3).
As an alternative to working directly with SARS-Cov-2, we sought to adapt our minimal, third-generation, lentiviral vector production system to produce recombinant HIV1 lentiviral vectors pseudotyped with the Spike (S) protein from multiple viral strains of SARS-CoV-2 (Wuhan Hu-1, Aus/VIC01, and S D614G). We term this SARS-CoV-2 pseudovirus, S-LV.
Our initial attempts to generate S-LV particles using transient transfection of 293T cells, grown in suspension, resulted in low yields (~1-2e5 TU/mL crude vector harvest) determined using our in-house, established hACE2 and hTMPRSS2 co-expressing indicator cells. We noted that S protein expression was modest at our routine post-transfection harvest time-points (48-72 hours depending on pseudotype), and hypothesised that delaying S-LV harvest to ~120 hours post-transfection would enhance S-LV particle yield. Gratifyingly, this simple manoeuvre boosted (P < 0.0001) S-LV yield by ~1 log to ~1-2e6 TU/mL. A further ~0.5-1 log increase in titre (P < 0.0001) was realised by removing an endoplasmic retention signal in the S protein by creating a 19 amino acid C-terminal deletion. Simple centrifugal concentration further increased S-LV titres.
Crucially, S-LV infectivity of our indicator cells was substantially (~80%) neutralised in vitro by two commercially available SARS-CoV-2 neutralising antibodies (nAbs; Sino Biologicals; P < 0.001). Thus, our indicator cells and S-LV preparation provides a functional neutralisation/inhibition assay to probe the ability of novel reagents (antibodies, small molecules, etc) to inhibit SARS-CoV-2 infection using a simple cell culture system that can under certain circumstances be used in a laboratory space categorised as BSL1. We are currently exploring the application of our platform anion exchange and tangential flow filtration approaches to further purify and concentrate S-LV particles to facilitate an in vivo neutralisation assay.
In conclusion, we now have a facile S-LV production method to facilitate the development of in vitro and in vivo neutralisation assays which can inform the development of SARS-CoV-2 therapeutics. The S-LV pseudovirus constitutes a useful reagent that can be produced and utilised in simple laboratory settings to progress COVID-19-related research.
Rapid prototyping and immunogenicity of SARS-CoV-2 DNA vaccine candidates formulated with the Fusogenix proteo-lipid vehicle delivery system
Douglas Brown, BSc, University of Alberta1, Arun Raturi, PhD, , John Lewis, PhD, , Roy Duncan, PhD, , Ping Wee, PhD, , Prakash Bhanduri, PhD, , Manoj Parmar, PhD, , Maryam Hejazi, PhD, , Liliya Grin, MSc, , Hector Vega, MSc, , Jennifer Gyoba, MSc, , Jailal Ablack, PhD, , Katia Carmine-Simmon, PhD, , Perrin Beatty, PhD, , Alyson Kelvin, PhD
University of Alberta1
There is an unmet need to develop a vaccine that provides durable immunity to SARS-CoV-2 in order to control its spread. Although robust neutralizing antibody responses have been measured in convalescent patients, research has indicated that immunity is short-lived, and reinfection is possible. To address these challenges, we initiated a rapid prototyping protocol to generate a panel of DNA vaccines (Covegenix) with the goal of stimulating potent neutralizing antibody and balanced cell-mediated responses while minimizing antibody-dependent enhancement. The DNA vaccine candidates carried SARS-CoV-2 sequences encoding full-length spike (FL-S) protein; wildtype, secreted, and point mutation versions of the receptor binding domain (RBD); and full-length nucleocapsid (FL-N) protein. Two different plasmid backbones and the genetic-encoded adjuvants RIG-I and CpG were also evaluated. The candidates were all formulated with the Fusogenix proteo-lipid vehicle (PLV) platform for delivery of the DNA vaccine into host cells. Fusogenix PLVs are formulated with a chimeric fusion associated small transmembrane (FAST) protein, which eliminates the requirement of toxic lipids for intracellular delivery while still maintaining efficient expression of DNA.
In total, we evaluated 24 potential COVID-19 DNA vaccine candidates for levels of neutralizing antibody and T cell immunity measured in a 1 or 2-dose vaccine candidate regimen over 28 days in preclinical mouse studies. FL-S stimulated the most robust neutralizing antibody titers, and the RIG-I and CpG adjuvants significantly improved responses. Secreted RBD produced the best response compared to all other RBD constructs and was able to achieve serum antibody concentrations as good as FL-S in the same DNA vector backbone. Neutralizing properties of the immunized mouse serum was determined using pseudotyped SARS-CoV-2. Serum from mice immunized with a single 25µg dose of FL-S and the RIG-I adjuvant produced a neutralizing titration curve similar to COVID-19 convalescent patients. Secreted RBD showed slightly lower titers but performed similar to FL-S from the same vector. Next, we examined cell mediated immunity with our lead candidates. FL-S+RIG-I showed a significant increase in antigen-specific IFN-γ producing T cells and activated CD8+ lymph node cells. We then examined the ability of FL-N at developing a durable pan-coronavirus vaccine, as N protein is highly conserved among beta-coronaviruses and leads to long lasting memory T cell populations. The FL-N candidates produced the strongest T-cell responses out of all vaccine candidates, with significant activated CD8+ cells in peripheral blood and spleen. Our screen has led to the development of an effective Covigenix DNA encoding both S and N antigens, important for generating a balanced humoral and cellular adaptive immune response against SARS-CoV-2 and other coronaviruses.
AAVCOVID: Immunogenicity In Mouse And NHP Of An AAV Gene-Based SARS-CoV-2 Spike Antigen Preventative Vaccine
Nerea Zabaleta, PhD, Mass Eye and Ear/Harvard Medical School1, Wenlong Dai, PhD, Maa Eye and Ear2, Urja Bhatt, Massachusetts Eye and Ear3, Julio Sanmiguel, Massachusetts Eye and Ear Infirmary4, Allison Cucalon, , Reynette Estelien, , Dawid Maciorowski, , Cheikh Diop, Massachusetts Eye and Ear Infirmary4, RUCHI CHAUHAN, PhD, GTVC5, Dan Li, , Abigail Sheridan, , Maya Kim, Mass Eye and Ear6, Kristofer Michalson, DVM, PhD, DACVP, , Cecilia Dyer, DVM, MS, DACLAM, , Jessica Chichester, PhD, University of Pennsylvania7, Marion McGlynn, MS, MBA, PMP, , Anna Honko, PhD, , Rebecca Johnson, PhD, , Nadia Storm, PhD, , Anthony Griffiths, PhD, , James Wilson, MD, PhD, University of Pennsylvania7, Mason Freeman, MD, , Luk Vandenberghe, PhD, Harvard8
Mass Eye and Ear/Harvard Medical School1
Maa Eye and Ear2
Massachusetts Eye and Ear3
Massachusetts Eye and Ear Infirmary4
GTVC5
Mass Eye and Ear6
University of Pennsylvania7
Harvard8
The clinical experience for AAV-based gene therapy treatments has steadily increased in the past decades, but the potential of AAV as a direct antigen immunization or vaccine platform has hardly been explored. AAV is known to be able to induce both B and T cell response against the capsid and the transgene. Generally, however, AAVs have a limited pro-inflammatory and even tolerogenic potential, which are both helpful for gene therapy, but undesirable for gene-based vaccines. AAVrh32.33 was previously generated as a chimera of two natural isolates from rhesus macaques that is uniquely capable of inducing functional anti-viral immunity upon IM injection in mouse and NHPs in a vaccine context. The goal of this study was to develop AAVrh32.33-based vaccines against SARS-CoV-2. We have designed and produced two clinically relevant vaccine candidates that consist of AAVrh32.33 carrying the stabilized full length SARS-CoV-2 Spike or the secreted S1 subunit of Spike, named AAVCOVID19-1 (AC1) and AAVCOVID19-3 (AC3), respectively. A single prime IM vaccination elicited high binding and neutralizing antibody responses in two mouse strains, BALB/c and C57BL/6 (n=5 mice/dose/gender/strain). Seroconversion was achieved for all animals treated at high dose (1x10
11 gc/mouse). Dose reduction modestly reduced both the rate of seroconversion and antibody levels, and delayed the kinetics of the response, in a gender-dependent manner. A robust correlation was found in binding and neutralizing titers, being pseudovirus neutralizing titers highest in AC1 high dose group (mean of 1:856). Antibody isotyping revealed a Th1/Th2 balanced response in animals treated with AC1 and a Th2 biased response in the AC3 groups. A pilot NHP study (n=2/candidate), intramuscularly dosed with 1x10
12 gc/NHP, revealed a kinetic difference between both candidates. AC3-injected animals raised binding antibodies from day 21 that continued to increase up to day 35. AC1 animals had detectable antibodies on day 35 and a continued increase afterwards. Live virus and pseudovirus neutralizing antibodies followed the same trend as the binding antibodies. In conclusion, AC1 and AC3 vaccines elicit high antibody responses in various preclinical models and showed quantitative and kinetic differences that were more evident in the NHPs. These candidates are being evaluated in long term studies to analyze the durability of response, in aged mouse studies and in prime-boost studies.
High Resolution Modeling of COVID-19 Disease Biology to Inform Gene and Cellular Therapies, and Vaccine Development
Renee Deehan, PhD, QuartzBio, Part of Precision for Medicine1, Josh Levy, MS, , Natalie Catlett, PhD, , Evan Gordon, BS, , Tobi Guennel, PhD, , Cliff Culver, BS, , Scott Marshall, PhD
QuartzBio, Part of Precision for Medicine1
Introduction
The current COVID-19 pandemic has created an arguably unprecedented urgency for identifying and developing treatments; and gene and cellular therapeutics represent modalities at the forefront of these efforts; as, for example, genetically based vaccines and viral antagonists, and serum transfers are currently being evaluated by translational and clinical programs. As we know, the host response to SARS-CoV-2 can vary based on comorbid condition status, age, and other factors; therefore it is critical for researchers to precisely understand the status of a patient’s manifestation of infection, but also response to therapy. Computational approaches applied to high-throughput biomarker measurements (e.g., RNA sequencing) represent a powerful way to model the biological state of a patient, treatment mechanism of action, and the translatability of pre-clinical model systems. Here we applied a prior knowledge-based, general artificial intelligence approach to identify the molecular mechanisms driving COVID-19 disease pathology in patients, and used that information to prioritize the best matched pre-clinical model for pre-clinical testing.
Methods
Publicly available RNAseq data from COVID-19 patient and non-infected lung biopsies was extracted from the Gene Expression Omnibus repository was used to identify transcripts that are significantly modulated in severe, active infections. Reverse causal inferencing, a method that leverages manually curated findings from thousands of peer-reviewed manuscripts, was applied to generate a high-resolution predictive model of molecular, cellular, and systemic functions that are dysregulated in disease. The same approach was taken to 1) evaluate two cell line models (NHBE and A549) for their ability to reflect the dysregulation inferred from COVID-19 patients, 2) characterize the impact of a therapeutic in ameliorating the dysregulated signaling caused by SARS-CoV-2 infection.
Results
Statistically significant mechanisms suggested by reverse causal inferencing supported a hyperactive innate immune response in COVID-19 patient characterized by increased Type 1 interferon signaling (increased IFNA, IFNG, IFNA13), increased toll-like receptor signaling (increased TLR7 and IRF5), decreased angiogenesis/remodeling (decreased FGF2, PDGF and ANGPT1), and potentially a dysfunctional adaptive immune response (decreased CD40L). When infected with SARS-CoV-2, the A549 cell line model better reflected COVID-19 pathogenesis compared than NHBE cells. Rituximab treatment of the A549 model mitigated dysregulated signaling induced by SARS-CoV-2 infection.
Conclusions
Reverse Causal Inferencing is a powerful computational modeling application to rapidly inform preclinical and clinical programs for gene and cell-based COVID-19 therapies about host response to infection, the utility of pre-clinical model systems, and the ability of a therapy to reverse the effects of SARS-CoV-2 infection.
Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs by a Simple Colorimetric Test
Bartolomeo Della Ventura, , Michele Cennamo, , Antonio Minopoli, , Raffaele Campanile, , Sergio Bolletti Censi, , Daniela Terracciano, , giuseppe Portella, Università Federico II napoli1
Università Federico II napoli1
Mass testing is fundamental to face the pandemic caused by the coronavirus SARS-CoV-2 and pushes the quest for fast, cheap but nevertheless reliable test to detect viral particles in order to identify infected subjects. We demonstrate that a colorimetric biosensor based on gold nanoparticle (AuNP) interaction induced by SARS-CoV-2 lends itself as an outstanding tool for detecting viral particles in nasal and throat swabs. The test is based on a highly scalable protocol making it suitable for realizing COVID-19 mass testing. A comparative analysis carried out on a total of 94 samples (45 positive and 49 negative) has shown both sensitivity and specificity higher that 95% with an area under the ROC curve of 0.98. The samples were collected in UMT (Universal Transport Medium). No additional treatments were performed; in particular, no RNA extraction and amplification was carried out. The test consisted in mixing the sample with properly functionalized AuNPs in colloidal solution. The presence of the viral particles (virions) induced nanoparticle aggregation that led to a redshift of the optical density in the extinction spectrum of the solution that was compared to the threshold cycle (Ct) of a Real Time-PCR (gold standard for detecting the presence of viruses) finding that the colorimetric method is able to detect very low viral load with a detection limit approaching that of RT-PCR. When the viral load was relatively high, i.e. with threshold Ct<15, the color change from red to purple was visible even by naked eye. Since the method is sensitive to the infecting viral particle rather than to its RNA, the achievements reported here open new perspective not only in the context of the current and possible future pandemics, but also in microbiology as the biosensor proves itself to be a powerful though simple tool for measuring the viral particle concentration.
Immunogenicity In Mice Of Nine AAV Spike-based COVID-19 Vaccine Candidates
Wenlong Dai, PhD, Maa Eye and Ear1, Nerea Zabaleta, PhD, Mass Eye and Ear/Harvard Medical School2, Urja Bhatt, Massachusetts Eye and Ear3, Reynette Estelien, , Dan Li, , Julio Sanmiguel, Massachusetts Eye and Ear Infirmary4, Abigail Sheridan, , Allison Cucalon, , Dawid Maciorowski, , Cheikh Diop, Massachusetts Eye and Ear Infirmary4, Maya Kim, Mass Eye and Ear5, RUCHI CHAUHAN, PhD, GTVC6, Luk Vandenberghe, PhD, Harvard7
Maa Eye and Ear1
Mass Eye and Ear/Harvard Medical School2
Massachusetts Eye and Ear3
Massachusetts Eye and Ear Infirmary4
Mass Eye and Ear5
GTVC6
Harvard7
The public health crisis of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains one of pandemic proportions. The development of a safe and efficient vaccine is therefore one of the highest priorities globally. Over the past decades, the adeno-associated virus vector (rAAV) platform has proven its safety and its potency as gene therapy agent. Its use as a preventative vaccine has been studied as well, however has received less attention. In the present study, we designed 9 COVID-19 genetic vaccine candidates with SARS-nCoV-2 Wuhan Spike antigen encoded as a transgene. The AAV2/rh32.33 capsid was used for gene delivery, based on the minimal seroprevalence, pro-inflammatory character following IM injection, and prior published reports in several preclinical vaccine settings. Here, AAV preparations were injected in Balb/c and C57BL/6 mice that were followed by regular phlebotomies to gauge the immunogenicity and vaccine potential after a single dose intramuscular (i.m.) immunization.
The antigenic profile of 9 alternate SARS-nCoV-2 Spike antigens are compared in an analogous AAV context. The design of the transgene antigens varies between full length, ectodomain, S1, and RBD only. In addition, some full length and ectodomain candidates have pre-fusion stabilized Proline mutations incorporated and the elimination of the S1/S2 furin cleavage site. Lastly, RBD only constructs have been designed as ss and scAAV.
Preliminary studies comparing the secreted monomeric S1 transgene with other candidates showed variable antibody responses and Th1/Th2 response types among candidates. The secreted monomeric RBD elicited lower binding and neutralizing antibody responses in C57BL/6 females and males. The WT full length showed similar binding levels as S1 at a low dose, but some animals had undetectable neutralizing antibodies (assay sensitivity <1:40). In contrast, the stabilized full length Spike showed slightly higher binding and neutralizing antibodies than S1 in both BALB/c and C57BL/6 females and males. Ongoing studies aim at comparing all 9 candidates side-by-side to assess binding and neutralizing antibody levels, Th1/Th2 responses and cellular responses.
These results demonstrate the potency and potential of AAVrh32.33 present us profound information of the immunogenicity and their immune response of AAV vectored vaccines, and provide us promising AAV vector based vaccine candidates against COVID-19 for further development.
Evaluation and optimization of SARS-CoV-2 pseudovirus
Hsu-Yu Chen, MS, University of Southern California1, Chun Huang, MS, University of Southern California1, Xiaoli Huang, , George Llewellyn, PhD, , Geoffrey Rogers, PhD, University of Southern California1, Lu Tian, PhD, , Ya-Wen Chen, PhD, , Paula Cannon, PhD, University of Southern California1
University of Southern California1
A major challenge for evaluating vaccine or therapeutic candidates against SARS-CoV-2 is the biohazardous nature of the live virus. Pseudoviruses based on lentiviral (LV), retroviral (RV), or vesicular stomatitis viral (VSV) particles that incorporate the SARS-CoV-2 Spike protein in their lipid envelopes are therefore a useful tool for certain aspects of SARS-CoV-2 research. While methods to produce and use such pseudoviruses have been reported for all three viral vector systems, limited studies have been done to compare their relative utility, or to optimize their production.
In this study, we evaluated and compared SARS-CoV-2 Spike pseudotyped LV, RV, and VSV vectors. We included the evaluation of both the native full-length Spike protein and a version with a cytoplasmic tail truncation to facilitate Spike incorporation. We also included the two dominant strains currently circulating, with residues D or G at position 614. Several strategies to enhance vector titer were evaluated, including different concentration methods. Vectors were examined for the level of Spike protein incorporation, titer on a reference cell line, ability to infect different target cell lines and a highly relevant lung organoid model, and susceptibility to neutralization by convalescent COVID serum.
Using a luciferase reporter gene, we found that VSV vectors supported 3-log higher titers when compared to RV vectors. Truncating Spike further enhanced vector titers more than 10-fold in VSV vectors, by facilitating incorporation into particles. Using tangential flow filtration to concentrate vectors resulted in a 3-fold higher yield than ultracentrifugation. When using the full-length Spike protein and VSV vectors the G614 strain showed enhanced infectivity on several target cell lines over D614, in agreement with recent reports. In addition, optimizing target cell density and the timing of any trypsin treatment further improved the reproducibility of vector titer measurements.
The results of this study provide a better understanding of the different properties and utilities of these three commonly used pseudovirus systems and suggest methods to enhance their production and facilitate their application in SARS-CoV-2 research.
Preclinical Immunogenicity Characterization of ARCT-021 SARS-CoV-2 Vaccine
Sean Sullivan, PhD, Arcturus Therapeutics, Inc.1, Eng Eong Ooi, BMBS PhD FRCPath, , Esther Gan, PhD, , Ruklanthi De Alwis, MPH PhD, , Shiwei Chen, PhD, , Daiki Matsuda, PhD, , Elizabeth Allen, , Jerel Vega, , Padmanabh Chivukula, PhD, , Rodrigo Yelin, PhD
Arcturus Therapeutics, Inc.1
ARCT-021 is a self-replicating RNA based vaccine developed to illicit protective immunity against SARS-CoV-2 infections. The RNA encodes for a viral replicase set of proteins that have been optimized to maximize expression of the SARS-CoV-2 spike glycoprotein (Sgp). The RNA is packaged in a cationic lipid formulation that protects the RNA from being degraded prior to cell entry and facilitates uptake and intracellular release into the cytosol. Mouse immunogenicity studies were conducted, and results were compared to an mRNA expressing the full length spike glycoprotein. The mRNA was formulated with the exact same lipid formulation as the self-replicating Sgp RNA, both lipid formulated RNAs having very similar physical/chemical characteristics. In Balb/c mice RNA dose dependent production of anti-Sgp IgG was higher and persisted longer with ARCT-021 than with Sgp mRNA vaccinations. Neutralizing antibody titers for ARCT-021 significantly increased at all timepoints tested whereas no neutralizing antibody titers were observed after single vaccination of Sgp mRNA. Vaccination end point titers showed equivalent titers for S1, S2 and RBD domains of the full length spike.
[SW1]
Cell mediated immune responses were evaluated in C57BL/6 mice by ELISPOT Sgp specific T lymphocytes and for Th1 and Th2 responses by intracellular cytokine staining. ARCT-021 produced an RNA dose dependent increase in CD8+IFN-γ+T lymphocytes whereas the % CD8+ IFN-γ+ T lymphocytes were less, and no RNA dose dependent increase was observed. ELISPOT analysis identified Sgp specific T lymphocyte responses to the RBD and S1-NTD domains of the Sgp. ARCT-021 vaccination produced a ≥ 4 fold higher % of positive T lymphocytes than Sgp mRNA. Th1 and Th2 responses were measured by assessing the %CD4+ IFN-γ+/CD4+IL-4 T helper lymphocyte ratio. The results supported a Th1 mediated immune response that was not skewed by a Th2 response. The Th1 mediated immune response was further supported by assaying the anti-Sgp IgG2a/IgG1 ratio in both Balb/c and C57BL/6 mice. A low RNA dose showed that ARCT-021 and Sgp mRNA had ratios >1, indicating a Th1 immune response. However, the anti-IgG2A/IgG1 ratios for higher RNA doses, ARCT-021 were between 8 and 10 [SW2] whereas the ratios for the Sgp mRNA vaccination were <1. These results show that the ARCT-021 SARS-CoV-2 vaccine elicits a robust humoral and cell mediated immune response after a single vaccination.
Cellular Immuno-Therapy for COVID-19 induced Acute Respiratory Distress Syndrome: Design of the CIRCA-19 Trial
Saad Khan, PhD, Ottawa Hospital Research Institute1, Dean Fergusson, PhD, , Manoj Lalu, MD, PhD, , David Courtman, PhD, , Mohamad Sobh, PharmD, , Irene Watpool, NP, , Josee Champagne, Msc, , Michael Jamieson, PhD, , Bernard Thebaud, MD, PhD, , Duncan Stewart, MD, PhD, Ottawa Hospital Research Institute1, Shane English, MD, PhD
Ottawa Hospital Research Institute1
The number of patients with the novel COVID-19 disease continues to rise world-wide. Approximately 20% of patients require hospitalization, and up to a quarter of these need care in the intensive care unit, mainly due to acute respiratory distress syndrome (ARDS). Mesenchymal stromal cells (MSCs) have been shown to reduce acute lung injury and improve ARDS in preclinical and clinical trials, including small, nonrandomized studies in COVID-19 patients. However, adequately designed randomized and appropriate controlled trials are needed to establish the safety and efficacy of MSCs for COVID-19 related ARDS. The Cellular Immuno-Therapy for COVID-19 related ARDS (CIRCA-19, NCT04400032) consists of 2 sequential trials: a phase 1 Vanguard trial and a phase 2 multicenter randomized trial (RCT).
The Vanguard trial is an open label dose escalation trial using a 3+3+3 design to determine the safety, and maximum feasible tolerated dose (MFTD) of intravenously (IV) delivered UC-MSCs. Up to 9 patients will be included; each receiving repeated unit doses of umbilical cord derived-MSCs (UC-MSCs) over 3 consecutive days (24±4 hours apart) according to the following dose-escalation schedule with 3 patients per dose panel: Panel 1: 25 million cells/unit dose (cumulative dose: 75 million MSCs); Panel 2: 50 million cells/unit dose (cumulative dose: 150 million MSCs); Panel 3: up to 90 million cells/unit dose (cumulative dose: 270 million MSCs). The Vanguard trial will provide valuable experience in identifying and overcoming the logistical constraints involved in delivering cell therapy products in the ICU during a pandemic including physician participation, patient identification and enrolment, and product distribution and administration. The phase 2 trial is a multi-center 2:1 randomized placebo-controlled study designed to evaluate efficacy (mechanical ventilator-free days) using UC-MSCs and will include 54 patients (36 UC-MSCs and 18 placebo). Participants allocated to UC-MSC intervention will receive 3 daily MSCs doses corresponding to the MFTD informed by the Vanguard trial.
UC-MSCs in CIRCA-19 are isolated from cords of healthy term pregnancies delivered by C-sections, cords are mechanically and enzymatically digested, and UC-MSCs are propagated in xeno-free conditions for 2 weeks prior to cryopreservation in a cord specific cell bank. Cell banks are validated and tested for freedom from adventitious agents, viability, MSC identity, and surrogate potency. To produce the MSC drug product, UC-MSCs from these cell banks are cultured for 48 to 96 h and provided as 2.5 x 106 UC-MSCs/mL suspended in PlasmaLyte A (PLA) containing 5% Human Albumin.
Participants will be eligible for inclusion if they meet all of the following criteria: 1) Age ≥ 18 years, 2) confirmed SARS-CoV-2 infection, 3) On invasive mechanical ventilation ≤48h, and 4) ARDS as per the Berlin criteria. The phase 1 Vanguard trial is now actively enrolling patients and details about accrual and tolerability of MSC therapy for COVID-19 ARDS will be available at time of presentation.
SARS-COV2 induced Neuroinflammation: A conduit to COVID-19 associated neuropathology.
Erin Clough, BS, University at Buffalo Dept of Medicine1, Lee Chaves, PhD, , Joseph Inigo, MS, , Dhyan Chandra, PhD, , Ravikumar Aalinkeel, PhD, , Stanley Schwartz, Md, PhD, , Jessica Reynolds, PhD, University at Buffalo2
University at Buffalo Dept of Medicine1
University at Buffalo2
Emerging Clinical data from the current pandemic suggests that ~40% of the patients with COVID-19 developed neurological symptoms attributed to viral encephalitis, resulting in neuro-inflammation, neuronal damage and severe hypoxia. The Neurological damage in COVID patients is attributed to the cytokine storm syndrome (CSS) which is characterized by systemic symptoms and signs derived from a massive and uncontrolled inflammatory response caused by pro- and anti-inflammatory cytokine dysregulation. Increased levels of pro-inflammatory cytokines such as interleukin (IL)-6, tumor necrosis factor (TNF)-alpha, IL-8, IL-10, were reported among fatal COVID-19 cases, indicative of hypercytokinemia, or CSS and may underlie encephalopathy. Angiotensin-converting enzyme 2 (ACE2 2) is the key entry receptor for SARS-CoV-2 which invades host cells in humans via binding to the spike protein of the virus and limited information is available on the distribution of ACE2 in neuronal cells such as normal human astrocytes ( NHA), brain microvascular endothelial cells (HBMVEC), and microglia(hμglia). We speculate that SARS-CoV-2 via the ACE2 receptor present on CNS cells exacerbates, neuroinvasion and neuroinflammation which increases anaerobic metabolism in the mitochondria of CNS cells resulting in mitochondrial dysfunction, hypoxia, increased oxidative stress and consequently neuronal death. Therefore, using real time PCR, Immunofluorescence staining, ROS activity assay cytokine arrays and Seahorse XFe96 Flux analyzer, the goal of our study was to examine, 1) the expression of ACE-2 on cells of the CNS cells, 2) the effect of SARS-COV-2 Spike protein on a) expression of key proinflammatory cytokines, IL-1β tumor necrosis factor (TNF)-α, MCP-1, IL-6, IL-8, IL-10; b) hypoxia inducible factors (HIF-1 & HIF-2); c) Reactive oxygen species ( ROS); and d) mitochondrial biogenesis in human microglial cells which are the major immune cells of the CNS. Our results show significantly increased expression of ACE-2 receptors, IL-1β, TNF-α, MCP-1, IL-6, IL-8, IL-10; HIF-1 and HIF-2 and ROS in SARS- COV2 spike protein treated NHA, HBMVEC and microglia as compared to untreated controls. Additionally, we also observed an increased Oxygen consumption rate (OCR) in microglial cells treated with SARS- COV2 spike protein. These data suggests that SARS- COV2 induces a significant inflammatory response, increased oxidative stress and a hypoxic milieu as indicated by increased expression of HIF’s and increased OCR all of which contribute to neuroinflammation and associated neuropathology of an encephalitic coronavirus infection and thus anti-cytokine based therapeutics may be effective in treating patients with COVID- 19 associated neurological disease.
Rapid Production Of Sars-Cov-2 Receptor Binding Domain By Viral And Non-Viral Expression Approaches For Pre-Clinical Evaluation
Omar Farnós, PhD, Department of Bioengineering, McGill University1, Alina Venereo Sanchez, PhD, McGill University2, Xingge Xu, , Cindy Chan, McGill3, Shantoshini Dash, McGill University2, Hanan Chaabane, , Denis Leclerc, Professor, , Amine Kamen, Professor, McGill University2
Department of Bioengineering, McGill University1
McGill University2
McGill3
Many different types of COVID-19 vaccine candidates are currently in development including vaccines based on replicating and non-replicating viral vectors, DNA, mRNA, subunit proteins, and virus-like particles (VLP) as well as inactivated and attenuated vaccines. Recombinant vaccines have reached important milestones in terms of safety and efficacy over the last decades. However, given the current knowledge of SARS-CoV-2 and the response of the human immune system to the viral infection, it is difficult to predict which class of vaccine will provide the best protection against the virus. It is therefore legitimate and strategically accurate to invest in the development of multiple vaccination platforms which will contribute to building long lasting capacities. In this work, various genetic constructions for different SARS-CoV-2 receptor binding domain (RBD) expression strategies have been developed. An extended RBD recombinant protein fragment for multipurpose clinical and vaccine research applications has been produced and purified following plasmid transient transfection or adenoviral infection of HEK293 cells cultured in suspension and serum-free medium. Characterization of the RBD expression in HEK293SF cells in 3L controlled bioreactors following these strategies led to high yield production of the highly purified RBD product with the appropriate conformational requirements for a variety of research and applied purposes. The extended RBD coding region was also inserted in the corresponding plasmid constructions for the development of simple and scalable manufacturing processes here described of human adenovirus type 5 vectors and adeno-associated virus (AAV) serotype 9, aimed for different vaccination approaches. The viral vectors were also produced in HEK293 cells cultured in suspension and serum-free medium in shake-flaks or 3L bioreactors, and were subjected to upstream and downstream process optimizations and subsequent characterization. Overall, this work set additional bases and provides important tools for the development of prophylactics and therapeutics against SARS-CoV-2.
LAMP-BEAC: Detection of SARS-CoV-2 RNA Using RT-LAMP and Molecular Beacons
Scott Sherrill-Mix, University of Pennsylvania1, Young Hwang, , Aoife Roche, , Susan Weiss, , Yize Li, , Jevon Graham-Wooten, , Louis Taylor, , Ronald Collman, , Gregory Van Duyne, , Frederic Bushman, PhD, University of Pennsylvania School of Med2
University of Pennsylvania1
University of Pennsylvania School of Med2
SARS-CoV-2 has caused a global pandemic, resulting in the need for rapid assays to allow diagnosis and prevention of transmission. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) provides a gold standard assay for SARS-CoV-2 RNA, but tests are expensive and supply chains are potentially fragile, motivating interest in additional assay methods. Reverse transcription and Loop Mediated Isothermal Amplification (RT-LAMP) provides a method that uses alternative and often cheaper reagents without the need for thermocyclers. The presence of SARS-CoV-2 RNA is typically detected using dyes to report bulk amplification of DNA; however a common artifact is nonspecific DNA amplification, complicating detection. Here we describe the design and testing of molecular beacons, which allow sequence-specific detection of SARS-CoV-2 genomes with improved discrimination in simple reaction mixtures. We also show how beacons with different fluorescent labels can allow convenient multiplex detection of several amplicons in “single pot” reactions.
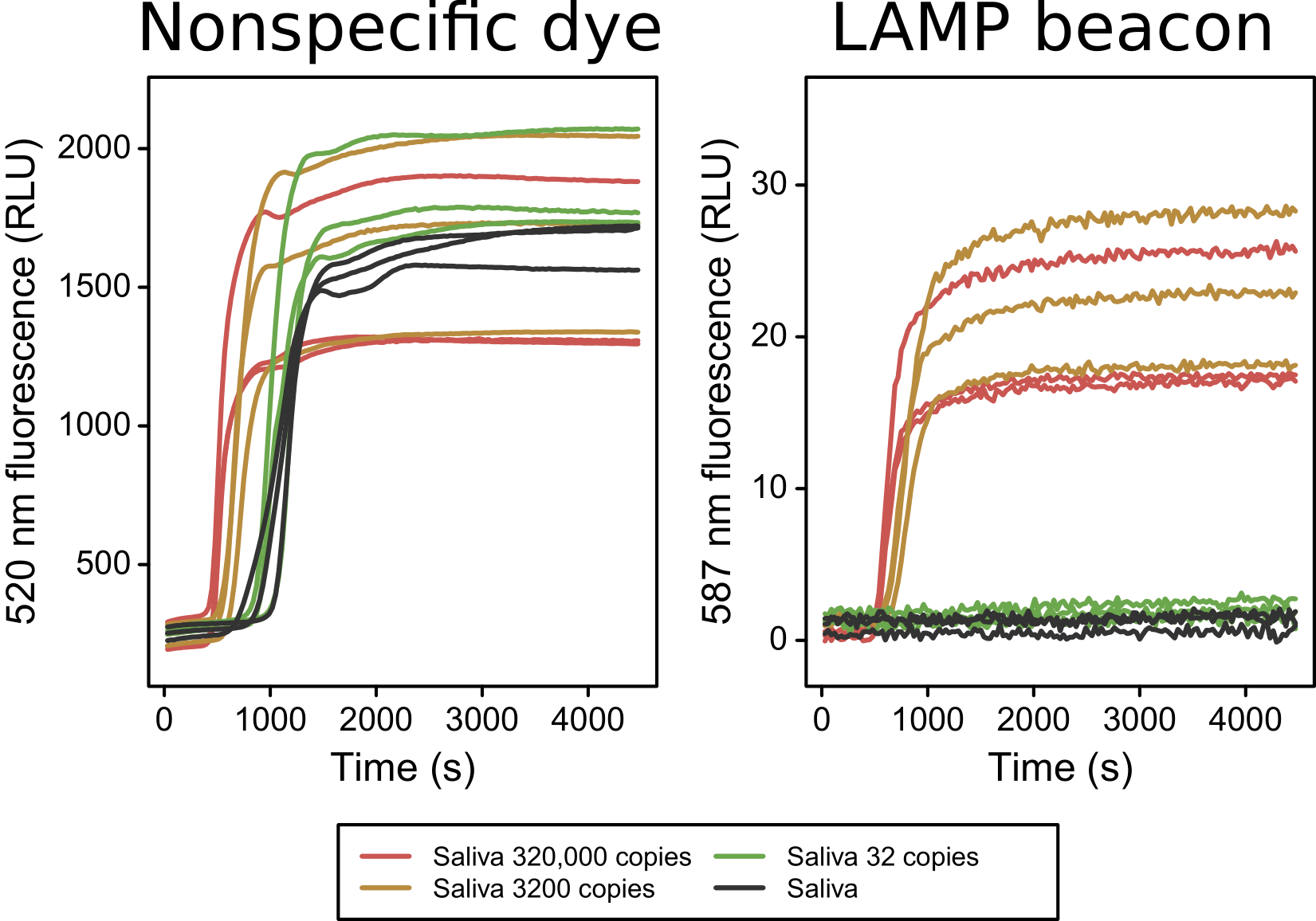
SARS-CoV-2 Spike Glycoprotein Activation in Cardiomyocytes: Mechanism and Pathological Consequences
Chanakha Navaratnarajah, PhD, Mayo Clinic1, Alison Barkhymer, , David Pease, , Daniel Clemens, , Dan Ye, PhD, , Chang Sung Kim, PhD, , Ryan Donohue, PhD, , Timothy Nelson, MD/PhD, , Michael Ackerman, MD/PhD, , Roberto Cattaneo, PhD, Mayo Clinic1, Jay Schneider, PhD
Mayo Clinic1
Overshadowed initially by the acute respiratory distress syndrome, COVID-19 myocarditis is now a major public health concern. We seek to understand the mechanisms of cardiac pathophysiology induced by SARS-CoV-2 infection. As with all coronaviruses, SARS-CoV-2 cell entry depends on its spike (S) glycoprotein, which mediates both binding to the ACE2 receptor and fusion of membranes. To fuse membranes, the S protein needs to be proteolytically activated. In airway epithelial cells, furin cleaves it at a multibasic cleavage site not found in closely related coronaviruses. However, proteolytic activation of the SARS-CoV-2 S protein is incompletely understood: in addition to furin cleavage at the S1/S2 site, it depends on cleavage by either the cell surface serine protease TMPRSS2, and/or endosomal cathepsins, at another site named S2’, located directly upstream of the hydrophobic peptide that inserts in the target membrane. We seek to test the hypothesis that furin cleavage may make this S2’ cleavage site more accessible, poising the S protein for mediating cell-cell fusion. We operated initially with Vero cells, which as cardiomyocytes do not express TMPRSS2. Using a furin inhibitor, we show that cell-cell fusion is furin-dependent. We also generated mutants of the S1/S2 cleavage site, and show that cleavage can be blocked by mutating the furin recognition sequence R-X-X-R. Importantly, this block can be reversed by overexpressing TMPRSS2 in Vero cells. However, while either furin or TMPRSS2 is sufficient to drive cell-cell fusion in Vero cells, presence of both proteases results in more extensive syncytium formation. We are mutating the S2’ TMPRSS2 recognition sequence to further characterize the role of the two protease cleavage sites, while also extending this work to human cardiomyocytes (hCMs) derived from induced pluripotent stem cells. In beating hCMs, expression of SARS-CoV-2 S-protein mediates production of giant multinucleated cardiomyotubes through cell-cell fusion. Cardiomyocytes containing hundreds of nuclei demonstrate profound functional derangements featuring pathological action potentials, Ca2+ sparks and tsunami-like Ca2+ fluxes. A furin inhibitor blocks SARS-CoV-2 S-protein driven cell-cell fusion of hCMs. With reports that many of the most severely affected COVID-19 patients suffer myocardial injury, we seek to use our model hCM system to elucidate some of the molecular mechanisms that underlie this pathophysiology.
Characterization of COVID-19 Gene Delivery Vectors by Charge Detection Mass Spectrometry
Benjamin Draper, PhD, Megadalton Solutions1, Martin Jarrold, PhD
Megadalton Solutions1
This work describes proprietary technology capable of directly measuring the mass and exact charge distributions of COVID-19 therapeutic targets. Mass spectrometry is one of the most powerful analytical techniques for characterization of biological molecules. However, most therapeutic approaches being investigated to date involve complexes falling outside the upper limit of conventional mass spectrometers (~1 MDa). We will provide a description of charge detection mass spectrometry (CDMS), and highlight how we are able to provide rapid and robust measurements of adenovirus and adeno-associated virus vectors. CDMS provides unmatched insight into the purity and content of gene therapy vectors. Measurements by CDMS require small quantities of material and can characterize lot-to-lot variability in the rapid scale-up of a widely administered vaccine. Such characterization will be invaluable to ensure immediate and long-term safety of a vaccine.
Figure 1. A) CDMS mass histograms of AAV control vectors produced insect cells. The vertical black dashed lines correspond to the expected mass based on the genome length packaged. The vertical green dashed lines correspond to the expected mass based on the packaging of two intact genomes, and the red vertical line represents the expected mass for a fully filled capsid. In each case the mass is greater and broader than expected indicating the presence of co-packaged fragment DNA along with the gene of interest. B) This co-packaging is confirmed through a pretreatment to the AAV capsids prior to measurement by CDMS where incubation at different temperatures is seen to eliminate co-packaging. This trend holds true for all genome lengths, and his highlighted by examining the CAG-GFP control vector. The mass of the peak attributed to the CAG-GFP shifts to the lower mass and sharpens indicating the presence of a single long strand with very little co-packaged content.
Figure 2. A) CDMS mass histograms of adenovirus control vectors. CDMS is able to measure the contents and quality of intact adenovirus vectors while also detecting incomplete and defective particles. Here we show three separate adenovirus control vectors with varied genome length. Adenovirus has shown great promise as a vaccine candidate for COVID-19 and the ability to quickly measure lot-to-lot variations and quality of adenovirus preparations could prove critical to rapidly scaling manufacturing of a vaccine.
Improved Spike Glycoproteins to Pseuodtype Lentiviral Vectors
Paul Ayoub, University of California, Los Angeles1, Roger Hollis, PhD, , Lili Yang, PhD, , Donald Kohn, MD, Unversity of California, Los Angeles2
University of California, Los Angeles1
Unversity of California, Los Angeles2
Introduction:
The spike (S) glycoprotein of SARS-Cov-2 facilitates viral entry into target cells through the cell surface receptor angiotensin-converting enzyme 2 (ACE2). Third generation HIV-1 lentiviral vectors can be pseudotyped to replace the envelope protein of the virus and thereby either limit or expand the target cell population. We aim to generate A pseudotyped lentiviral vector enveloped by a modified S glycoprotein of SARS-Cov-2 capable of infecting ACE2 expressing cells more efficiently than that of wildtype S glycoprotein.
Methods:
We generated a 3rd generation HIV-1 pseudotyped lentiviral vector enveloped by Spike glycoprotein with an mCitrine transgene cassette. Prior research has suggested that multiple viral envelope proteins can pseudotype lentiviral vectors more effectively, such as that of influenza hemagglutinin (HA) and the murine leukemia virus cytoplasmic tail. As a result, many variables were considered when designing these new glycoproteins including 1) cytoplasmic tail modifications 2) mutations and 3) codon optimizations. Spike variants were packaged into lentiviruses with encoding for an mCitrine transgene cassette. ACE2 expressing cells were transduced with the spike pseudotyped lentiviral particles. The effectiveness of each glycoprotein variant was measured for its infectious particles, its efficiency of particle formation, and its particle functionality. These modifications led us to generate a candidate vector with infectivity and expression than previous iterations.
Results:
Using our pseudotyped lentiviruses driving an mCitrine reporter, we demonstrated a variant capable of achieving 10-fold higher expression in ACE2 expressing cells than wildtype spike glycoprotein. Calu-3 and ACE-293T cells were transduced with each pseudotyped vector. Tranducing units per mL (TU/mL) were measured by ddPCR to quantify the infectious particles of each variant. Furthermore, GFP percentage and MFI was measured by FACS to quantify vector functionality and expression. Finally, spike protein and p24 was measured by ELISA to quantify the efficiency of particle formations and to rule out bald particles.
Developing IL-27-based gene delivery for protecting and treating COVID-19 Lung Disease
Grace Mulia, BS, Purdue University1, Janelle Salameh, , Richard Kuhn
Purdue University1
COVID-19 has impacted millions of people worldwide, with over 20 million positive cases globally and over 700,000 deaths and counting. As preventative vaccines and effective antivirals remain unavailable, host-directed, preferably targeted therapeutics employing immunomodulatory agents must be explored for systemic delivery. Many ongoing efforts are getting the research community closer to discovering safe and efficacious treatments and/or vaccines to combat this devastating pandemic.The immune response tips radically off-balance in patients with severe COVID-19 and current therapies are unable to stop the lung destruction and mortality. The risk factors are incompletely understood and contributing to the mortality are serious systemic effects on the cardiovascular and immune systems. Additionally, COVID-19 patients are observed to have an imbalance in cytokine expression, with the upregulation of IL-6, IL-8, IL-1, TNF-which correlates with the severity of disease outcome and organ damage. Therefore, perhaps a therapeutic that can immunomodulate and rebalance cytokine profiles while possibly reducing the severity of the infection is being investigated by our group. Interleukin-27 (IL-27), a versatile cytokine that is involved in multiple inflammatory signaling pathways, shows promise in its ability of rebalancing the lung or other tissue microenvironments. We hypothesize that the multifunctional IL-27 biologic agent can rebalance the COVID-19 inflammatory microenvironment in lung and other tissue culture models. We plan to examine the potential for IL-27 gene delivery approaches in inducing expression of antiviral genes and in restoring the cytokine imbalancein vitro. Our study may help determine the potential of these therapeutics in restoring immune balance which could lead to novel approaches for reducing the progression of severity and mortality in COVID-19.
Synthetic DNA-delivery of a SARS-CoV-2 Neutralizing Antibody with Durable In Vivo Expression and Neutralizing Activity
Elizabeth Parzych, PhD, , Mamadou Bah, , Nick Tursi, , Ebony Gary, PhD, The Wistar Institute1, David Weiner, PhD, The Wistar Institute1, Dan Kulp, PhD, , Emma Reuschel, PhD
The Wistar Institute1
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has become a priority target for vaccine, drug and therapeutic development. Recombinant monoclonal antibodies (mAbs) are important biologics but present challenges in terms of rapid, large-scale production and widespread deployment in response to a global pandemic. DNA-encoded monoclonal antibodies (DMAb) represent an alternative development and delivery system for the in vivo production of antibody-based biologics. The deployment of this technology against a diverse set of infectious diseases, including emerging pathogens such as Zika and Ebola viruses, have been described. Here, we designed and modified in silico a human SARS-CoV-2 DMAb based on a recently reported neutralizing mAb clone (Pinto et al. 2020. Nature. 583: 290-295) that targets the receptor binding domain (RBD) of the SARS-CoV-2 spike protein. Variable domain sequences were modified, optimized and cloned into a previously studied expression vector. In vitro transfection of these plasmid constructs resulted in the assembly and secretion of IgG that exhibited potent target binding and inhibitory activity in a pseudoviral neutralization assay. Constructs were then delivered in vivo via electroporation, resulting rapid seroconversion and robust expression levels exceeding 30 ug/ml. Significant expression was detected in the sera for at least 2 months post-administration. Importantly, sera from DMAb-administered mice exhibited both target binding and significant inhibitory activity. These data demonstrate that the design, in vivo production and evaluation of neutralizing antibodies against SARS-CoV-2 can be readily achieved for potential clinical translation. This has promising implications for the further development of novel, antibody-based tools against new emerging infectious diseases using the DMAb platform.
Anti-Covid MicroRNA Therapy to Block the Expression of the Spike, Envelope, Membrane, and Nucleocapsid Genes of SARS-CoV-2
Martin Hicks, PhD, Monmouth University1, Flobater Gawargi
Monmouth University1
Emerging viral diseases have increased in recent decades. In December 2019, an epidemic with low respiratory infections emerged in Wuhan, China. The disease, Covid-19 was found to be caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). As of August 14, 2020, WHO has confirmed 20,950,402 global cases and 760,213 deaths worldwide, 167,253 in the USA. To date, there are no vaccines nor specific therapeutic drugs against SARS-CoV-2. From advances in biotechnology, the genome and structure of SARS-CoV-2 is known. Three proteins are anchored in the viral envelope, Spike (S), Envelope (E), and Membrane (M), which is linked to the Nucleocapsid (N) protein connecting to the viral RNA genome. Our lab is developing an innovative therapy that delivers multiple therapeutic microRNAs to simultaneously block the expression of these distinct viral proteins. In the current work, we propose an anti-Covid microRNA therapy designed to degrade each of the mRNA transcripts of these critical genes, stopping viral assembly, and reducing the severity of infection. The design of the anti-Covid microRNAs 1) mimics microRNA cluster 17-92 structural stability, 2) forms guide-RNA substrates for the RNA induced silencing complex, and 3) are complementary to specific regions of the SARS-CoV-2 RNA genome without off-targets effects in the human genome. Twenty-one microRNA sequences were designed to target the S gene, six for N, two for M, and one for E. In the first phase, six microRNA sequences have been cloned into microRNA-therapy vector. Each of the genes, S, E, M and N will be transfected into our tissue culture model to measure the efficacy of the anti-Covid microRNA therapy to down-regulate the expression of the viral genes. To advance our therapeutic strategy, we will be analyzing the secondary structure and stability of the anti-Covid miRNA using selective 2’ hydroxyl acylation and primer extension followed by mutational profiling (SHAPE-MaP). In the Fall Semester 2020, we are introducing Covid-19 therapeutics into our Genetics course. Based on our strategy, we are engaging undergraduate students to individually design a therapy to target each of the SARS-CoV-2 viral genes and subsequently test their efficacy to knockdown the expression of the viral proteins with the potential to optimize our therapeutic strategy while educating the next generation of scientist.






























